
Advances in Pediatric Research
Open Access
ISSN: 2385-4529

ISSN: 2385-4529
Research Article - (2024)Volume 11, Issue 2
Background: One hundred fifty-seven preterm infants who were enrolled in the study were hospitalized between 2012 and 2014 at the University Hospital of Amiens-Picardie. Only 28 (17.8%) of the children who had a secondary coagulase-negative staphylococcal bacteremia with positive stool cultures were included in this study.
Objectives: This investigation sought to examine the rate of intestinal bacterial translocation associated with these infections.
Methods: In the context of this study, blood and stool cultures were performed. MALDI-TOF MS was used to examine all isolates of Staphylococcus spp. Genotyping and antibiotic susceptibility were also done.
Results: Antibiotic susceptibility revealed sixteen resistance patterns in blood and stool. Ten of the coagulase-negative Staphylococcus strains isolated from blood samples showed an R pattern e (35.7%) and eleven of the coagulase-negative Staphylococcus strains isolated from stool samples showed an R pattern e (39.2%). 53.5% of cases in blood culture results were similar to stool culture results and in 46.5% of cases they were different. Fifteen different bacteria had three different patterns: ERIC-2 (A, B and C) and RAPD-PCR (D, E and F). ERIC-2 patterns comprised A (S. epidermidis isolates); B (S. haemolyticus isolates) and C (unidentified coagulase-negative Staphylococcus isolates). RAPD patterns consisted of D (unidentified coagulase-negative Staphylococcus isolates); E (S. haemolyticus isolates) and F (S. epidermidis isolates).
Conclusion: Bacterial translocation from the gastrointestinal tract may have been the cause of coagulase-negative staphylococcal bacteremia in hospitalized preterm infants.
Bacterial translocation; Coagulase-negative Staphylococcus; Blood culture; Stool culture; Preterm infants; Molecular typing
AC-FMS: Antibiogram Committee-French Microbiology Society; ARDS: Acute Respiratory Distress Syndrome; BT: Bacterial Translocation; BW: Birth Weight; CIs: Confidence Intervals; CLDN: Claudine; CoNS: Coagulase Negative Staphylococci; CRP: C-reactive Protein; ERIC-PCR: Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction; EUCAST: European Committee on Antimicrobial Susceptibility Testing; GA: Gestational Age; GIT: Gastrointestinal Tract; JAM: Junctional Adhesin Molecule; MALDI-TOF-MS: Matrix-Assisted-Laser Desorption Ionization Time of Flight Mass Spectrometry; MLNs: Mesenteric Lymph Nodes; MODS: Multiple Organ Dysfunction Syndrome; MOF: Multiple Organ Failure; MSA: Mannitol Salt Agar; NF-κB: Nuclear Factor-Kappa; NI: Nosocomial Infection; NICUs: Neonatal Intensive-Care Units; OCLN: Occludin; PAMPs: Pathogen-Associated Molecular Patterns; PICUs: Pediatric Intensive Care Units; RAPD-PCR: Random Amplification of Polymorphic Deoxyribonucleic Acid (DNA)-Polymerase Chain Reaction (PCR); REP: Repetitive Extragenic Palindromic; RFs: Risk Factors; RR: Risk Relative; S: Staphylococcus; SD: Standard Deviation; SIRS: Systemic Inflammatory Response Syndrome; TLC: Total Leucocyte Count; TLR: Toll-like Receptors; TPN: Total Parenteral Nutrition; U: Unidentified; UHAP: University Hospital of Amiens-Picardie; VLBW: Very-Low-Birth-Weight; WG: Week Gestation; ZO: Zonula Occludens
Coagulase-Negative Staphylococci (CoNS), commonly referred to as opportunists, constitute one of the most significant nosocomial pathogens, causing a substantial deterioration in human life and health [1]. They have been identified as the primary factor responsible for late-onset sepsis in Neonatal Intensive-Care Units (NICUs). Several risk factors have been reported, including extreme prematurity and a Very-Low-Birth-Weight (VLBW), as well as the use of intravascular catheters, prosthetic device-related sepsis and prolonged parenteral nutrition [1,2]. Despite the consensus among published studies that CoNS exhibit a relatively low virulence and are not associated with significant morbidity. It has been reported that atypical outbreaks of persistent bacteremia have also been reported despite aggressive antibiotic therapy [3]. The process of adhering to prosthetic material and then making a biofilm matrix (slime) is complicated. The ica operon is an important element in this process, as it encodes the polysaccharide intercellular adhesin (icaA, icaD, icaB and icaC genes). The intercellular adhesion of bacteria and the multilayer accumulation of the biofilm, make them especially resistant to antimicrobial agents and immune functions of the host [4]. Though the relevance of the presence of the ica operon and the production of slime has already been studied in neonates. It’s not clear if the bacteria that cause more severe forms of CoNS sepsis use these factors [2].
Sepsis is a potentially fatal organ dysfunction that is correlated with a dysfunctional host response to infection [5,6]. It is a significant cause of mortality due to Multiple Organ Failure (MOF) in critically ill patients [7]. The development of gut barrier dysfunction and Bacterial Translocation (BT) in sepsis is influenced by both bacteria and host factors [8-11]. The intestinal bacteria or their products can through the epithelial barrier, is which can lead to many human diseases like inflammatory bowel diseases, obesity, cancers and postoperative complications. The mesentery is thought to be a key tissue site that help these “escaped” microbial signals become physiological and immune [12]. The Gastrointestinal Tract (GIT) is a conduit for invasive infections in critically ill individuals, owing to an increased in BT associated with compromised barriers. Intestinal bacteria can be disseminated systemically via BT, moving through the Mesenteric Lymph Nodes (MLNs) or activating the gut immune system [13,14]. MOF is caused by activation of the immune system and the secretion of pro-inflammatory cytokines due to intestinal barrier and BT dysfunction [15,16].
There are many pathways, as shown in Figure 1, receptors and cells involved in BT from the intestinal lumen. Toxins like flagellin, endotoxins, exotoxins and other bacterial products can break tight junctions Junctional Adhesion Molecule (JAM); Occludin (OCLN); Zonula Occludens (ZO); Claudine (CLDN) and help paracellular translocation of bacteria between intestinal epithelial cells. Transcellular translocation of bacteria can occur through receptors such as intelectin (also known as lactoferrin (LF) receptor), the type III secretory system, Toll-Like Receptors (TLR), Lymphocyte Function associated Antigen-1 (LFA-1), lectin receptor β1 integrin and IgA which are present on M cells. The dissemination of the microbe through these cells can result from the bacterial uptake [16]. The development, maintenance and repair of an intestinal villus, is facilitated by involvement of commensal bacteria and paneth cells. The cells at the bottom of the crypts make four types of cells: enterocytes, goblet cells, paneth cells and intestinal neuroendocrine epithelia. These cells are involved in host defense against BT, as depicted in Figure 1. The significance of neuroendocrine epithelia in the defense of the intestinal villus by the host remains uncertain. Physical, biochemical and immunological factors play a role in the intestinal barrier dysfunction. Endotoxins and antigens are sent from the gut into the blood [17]. Sepsis is a condition in which bacteria and bacteria-associated products cause a systemic response. These reactions in the blood cause other symptoms in people who are very sick, like Acute Respiratory Distress Syndrome (ARDS) and Multiple Organ Dysfunction Syndrome (MODS) [5]. BT can cause an inflammatory response, called cytokine-mediated Systemic Inflammatory Response Syndrome (SIRS) and MODs [15]. The underlying mechanisms behind the emergence of SIRS and MODs are linked to bacterial mediators leaving the gut referred to as “danger particles” or Pathogen-Associated Molecular Patterns (PAMPs). PAMPs are believed to stimulate the innate immune system. Both viable bacteria and PAMPs can cause sepsis by acting on the immune system [18-21]. These factors increase the expression of Nuclear Factor Kappa light-chain enhancer of activated B cell (NF-κB)-associated inflammatory transcription factors and the excess production in inflammatory cytokines associated with end-organ damage [22,23]. The aim of this study was to evaluate the rate of intestinal BT and its role in causing sepsis in clinical conditions, as well as the correlation between CoNS isolated from blood cultures and stool cultures using molecular typing.
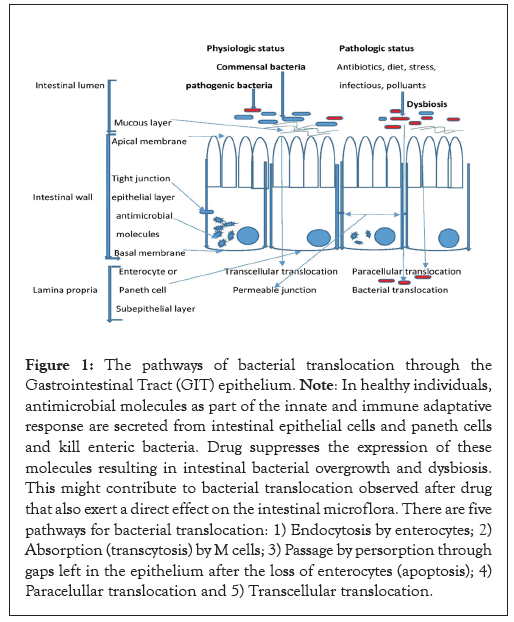
Figure 1: The pathways of bacterial translocation through the Gastrointestinal Tract (GIT) epithelium. Note: In healthy individuals, antimicrobial molecules as part of the innate and immune adaptative response are secreted from intestinal epithelial cells and paneth cells and kill enteric bacteria. Drug suppresses the expression of these molecules resulting in intestinal bacterial overgrowth and dysbiosis. This might contribute to bacterial translocation observed after drug that also exert a direct effect on the intestinal microflora. There are five pathways for bacterial translocation: 1) Endocytosis by enterocytes; 2) Absorption (transcytosis) by M cells; 3) Passage by persorption through gaps left in the epithelium after the loss of enterocytes (apoptosis); 4) Paracelullar translocation and 5) Transcellular translocation.
Study design and study population
This study is retrospective, non-interventional, that does not involve the human person and is conducted exclusively from the exploitation of existing medical data collected from electronic medical records. During the study period, a total of 157 neonates (with a life expectancy of less than 28 days) who were born prematurely (<37 Weeks of Gestation (WG)) and clinically suspected of neonatal sepsis were promptly admitted to the NICUs and Pediatric Intensive Care Units (PICUs) at the University Hospital of Amiens-Picardie (UHAP), Amiens (France) between 2012 and 2014. Blood culture was done for these neonates. All neonates who had a positive blood culture for CoNS and a concomitant positive stool culture were included in the study as potential cases of CoNS sepsis. All preterm infants who had a negative blood culture for CoNS, 129 (82.2%), who received antibiotics before being admitted to the NICUS or PICUs, as well as those who died before their blood was tested, were not included.
Data collection
All of the data was taken from electronic medical records. Patients’ demographics, underlying conditions and clinical and laboratory findings were all collected.
Diagnostic criteria for bacteremia
One positive blood culture was detected in the presence of suggestive clinical manifestations, including bradycardia, oxygen desaturation, increased respiratory requirements, Total Leukocyte Count (TLC) of >18,000/mm³, platelet count of >450,000/mm³, C-reactive Protein (CRP) of >10 mg/L and lactate of >1.65 mmol/L, temperature >38.3°C or <36°C.
Blood and stool samples
Blood collected for routine hematological, biochemical and microbiological examinations, including TLC, platelet count, CRP and serum lactate. During this study, all blood and stool cultures were performed for diagnostic purposes. Less than 0.5 mL of blood was inoculated into a bactec peds plus F bottle and incubated in a bactec TM Becton Dickinson (BD) instrument (BD diagnostic systems, spark, MD, USA). Subcultures of the initial blood culture broth were seeded on sheep blood (5%) Columbia agar and Mannitol Salt Agar (MSA) (Oxoid, France) and incubated aerobically at 35 ± 2°C for 24 hours. Anal or rectal samples were seeded on sheep blood (5%) Columbia agar and MSA and incubated aerobically at 35 ± 2°C for 48 hours.
Identification of isolates
After examination of all colonies, the identification of Staphylococcus spp. was determined. All isolates that tested negative for mannitol and bound coagulase (Pastorex Staph Plus-Bio-Rad, France) and positive for catalase and gram staining, were classified as CoNS.
MALDI-TOF-MS
Matrix-Assisted-Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS) (Brucker Daltonics, Bremen, Germany) was used to examine all Staphylococcus strains identified by routine tests, following a previously described procedure [24,25].
If there was a negative blood culture, the stool cultures and blood cultures were thrown away. On the contrary, when a Staphylococcus spp. was found in a patient’s blood sample, all the different colonies that were found in stool were identified using MALDI-TOF-MS.
Antibiotic susceptibility testing
The disk diffusion method was used to test isolates on Mueller-Hinton (MH) agar according to the zone size criteria recommended by the Antibiogram Committee of the French Microbiology Society (AC-FMS) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST)-2015 [26]. The antibiotics used were Kanamycin (K), Gentamicin (G), Tobramycin (T), Erythromycin (E), Lincomycin (L), Pristinamycin (PT), Rifampin (RIF), Ofloxacin (OFX), Vancomycin (VAN), Fusidic acid (FA), trimethoprim Sulfamethoxazole (SXT) and Fosfomycin (FOS). The susceptibilities to Benzylpenicillin (P) (disk loaded with 6 µg) and Cefoxitin (FOX) (disk loaded with 30 µg) were determined by the disk diffusion method on salted MH for incubation at 37°C for 24 hours.
DNA isolation and molecular typing
Following to the manufacturer’s instructions, total nucleic acid extraction was performed using the bioMérieux NucliSENS easy MAG platform (bioMérieux, Marcy l’Etoile, France). The ERIC-PCR procedure was followed in accordance with previous reports [27]. Extracted DNA (100 ng from each isolate) was amplified in a final volume of 50 µL of the ERI-C2 primer (5’-AAG-TAA-GTG-ACT-GGG-GTG-AGC-G-3’) and 5 µL of coralload PCR buffer. PCR cycling consisted of 94°C for 7 min followed by 45 cycles of 94°C for 1 min, 45°C for 1 min and 72°C for 2 min and 72°C for 7 min. The RAPD-PCR procedure was followed in accordance with previous reports [28]. The primer used was, 5’-GGT-TGG-GTG-AGA-ATT-GCA-CG-3’. Amplification reactions were carried out with a final volume of 50 µL containing 25 µL of Top Taq Master MIX (QIAGEN, Les Ulis, France), 1 µL of RAPD1 primer, 2 µL of DNA as template and 5 µL of coralload PCR buffer. The cycling conditions were 95°C for 3 min, followed by 35 cycles of 94°C for 1 min, 40°C for 1 min, 72°C for 2 min and 72°C for 10 min. After amplifications, PCR products were resolved by electrophoresis on 1.2% agarose gels at 90 V for 6 h, followed by ethidium bromide staining and were visualized under UV light. A photograph was also taken.
Risk Factors (RFs) for gastrointestinal BT in preterm infants with CoNS bacteremia
The following clinical, laboratory and treatment parameters were evaluated as potential RFs for BT. According to molecular typing, these RFs were divided into two groups: Group 1 included patients in whom BT was indicated and group 2 comprised patients did not need a BT.
Statistical analysis
We have computed the proportional Risk Relative (RR) of BT in preterm infants with and without documented BT. For quantitative variables, data are expressed as means ± Standard Deviation (SD) and for qualitative variables, as frequencies. We used fisher’ s exact test to figure out the RR of BT with associated 95% Confidence Intervals (CIs). Comparative analyses between documented and undocumented BT groups were performed using the wilcoxon-mann-whitney test for quantitative data and fisher’s exact test for qualitative data. The significance tests were all two-sided and a Probability (P) value of <0.05 was considered to indicate statistical significance.
Patient’s characteristics
This study analyses the characteristics of 28 preterm infants included in this study (mean ± SD, median and range). Gestational Age (GA) (weeks): 29.2 ± 3.3, 29 and 25-36; Birth Weight (BW) (gram): 1,288 ± 606.2, 1,075 and 592-2,800; age at onset of infection (days): 19.8 ± 21.5, 11 and 3-104; lactate (µmol/L): 38.2 ± 43.5, 23.3 and 3.0-43.5; CRP (mg/L): 3.28 ± 1.62, 5.15 and 0-178; TLC (/mm³): 26,775 ± 48,733.7, 15,400 and 5,000-271,000; platelets count (/mm³): 162,642.8 ± 98,180.5, 54,500 and 60,000-286,000; maternal age at birth (years): 29.7 ± 6.2, 29.5 and 19-45 (Tables 1 and 2).
| Patient No. | Gestational age *(WA) | Birth weight (g) | Age at onset of infection (days) | Gastrointestinal disorders | Lactates mmol/L | ***CRP mg/L | Leukocytes/mm³ | Platelets/mm³ |
|---|---|---|---|---|---|---|---|---|
| 1 | 26 | 710 | 48 | Yes | 7.2 | 0 | 9900 | 70000 |
| 2 | 25 | 950 | 47 | Yes | 7.3 | 5 | 9800 | 214000 |
| 3 | 36 | 2800 | 8 | Yes | 5.5 | 53 | 9900 | 230000 |
| 4 | 26 | 700 | 8 | Yes | 7 | 5.7 | 34200 | 173000 |
| 5 | 28 | 1200 | 30 | Yes | 6.9 | 40 | 19900 | 65000 |
| 6 | 29 | 1370 | 9 | Yes | 6.8 | 23 | 18400 | 200000 |
| 7 | 27 | 975 | 11 | Yes | 7.5 | 62 | 25100 | 85000 |
| 8 | 28 | 960 | 13 | Yes | 7.7 | 17 | 13400 | 103000 |
| 9 | 30 | 1060 | 7 | No | 3.5 | 0 | 12000 | 213000 |
| 10 | 31 | 1720 | 10 | No | 4.5 | 178 | 20500 | 70000 |
| 11 | 11 | 1730 | 10 | Yes | 5.8 | 84 | 19000 | 60000 |
| 12 | 27 | 986 | 19 | No | 4.6 | 3.4 | 11800 | 316000 |
| 13 | 25 | 700 | 28 | Yes | 6.5 | 0 | 11400 | 194000 |
| 14 | 35 | 2371 | 14 | Yes | 6.8 | 99 | 10500 | 136000 |
| 15 | 26 | 592 | 15 | Yes | 7.9 | 103 | 16100 | 72000 |
| 16 | 27 | 990 | 8 | Yes | 2.9 | 89 | 271000 | 12000 |
| 17 | 29 | 1330 | 3 | Yes | 3.4 | 58 | 37000 | 123000 |
| 18 | 29 | 755 | 6 | Yes | 4.2 | 0 | 27600 | 98000 |
| 19 | 32 | 1090 | 53 | Yes | 4.3 | 23.7 | 9500 | 298000 |
| 20 | 35 | 2400 | 6 | Yes | 5 | 41.5 | 20400 | 73000 |
| 21 | 36 | 2730 | 104 | Yes | 5.9 | 15 | 9100 | 199000 |
| 22 | 29 | 949 | 5 | Yes | 5.3 | 31 | 10700 | 232000 |
| 23 | 34 | 1340 | 24 | Yes | 3.5 | 45 | 14700 | 185000 |
| 24 | 30 | 1250 | 13 | Yes | 3.7 | 10 | 18200 | 447000 |
| 25 | 29 | 1320 | 5 | No | 4.1 | 0 | 5000 | 120000 |
| 26 | 28 | 1192 | 11 | Yes | 3 | 0 | 12000 | 286000 |
| 27 | 26 | 995 | 33 | Yes | 3.3 | 84 | 36900 | 75000 |
| 28 | 25 | 900 | 9 | No | 4 | 0 | 35700 | 205000 |
| Mean ± SD | 29.2 ± 3.38 | 1288 ± 606.2 | 19.8 ± 21.5 | Y: 82.2% | 38.2 ± 43.5 | 3.28 ± 1.62 | 26775 ± 48733.7 | 162642 ± 98180 |
| N: 17.8% | ||||||||
| Median | 29 | 1075 | 11 | - | 23.3 | 5.15 | 15400 | 54500 |
Note: WA: Weeks of Amenorrhea; ***CRP: C-Reactive Protein; SD: Standard Deviation.
Table 1: Characteristics of the 28 children with coagulase-negative Staphylococcus spp. included in the study.
| Gestational Age (GA-weeks) | Range | N (%) |
| Extremely preterm (<28) | 25-27 | 10 (35.7) |
| Very preterm (<32) | 21-31 | 12 (42.8) |
| Moderate preterm (<37) | 32-36 | 6 (21.5) |
| Total | - | 28 (100.0) |
| Birth Weight (BW-gram) | Range | N (%) |
| Extremely Low Birth Weight (ELBW)<1000 | 592-995 | 13 (46.5) |
| Very Low Birth Weight (VLBW)<1500 | 1060-1370 | 9 (32.2) |
| Low Birth Weight (LBW)<2500 | 1720-2400 | 4 (14.2) |
| Normal birth weight ≥ 2500 | 2730 and 2800 | 2 (7.1) |
| Total | - | 28 (100.0) |
Table 2: Distribution of 28 preterm infants according to their gestational age and their birth weight.
Blood culture and stool culture results
S. haemolyticus and S. epidermidis were isolated from 39.3% and 17.8% of blood cultures from these patients, respectively, 35.7% of blood cultures showed unidentified Coagulase-negative Staphylococcus spp. (uCoNS). S. haemolyticus and S. epidermidis were isolated from 42.8% and 17.8% of stool cultures respectively and uCoNS were isolated from 39.4% of stool cultures, which is shown in Table 3.
| Coagulase-negative Staphylococcus spp. | Blood culture N (%) | Stool culture N (%) | Total |
|---|---|---|---|
| *S. epidermidis | 5 (17.8) | 5 (17.8) | 10 (17.8) |
| S. haemolyticus | 11 (39.3) | 12 (42.8) | 23 (41.0) |
| S. warneri | 1 (3.6) | 0 | 1 (1.8) |
| S. capitis | 1 (3.6) | 0 | 1 (1.8) |
| **uCoNS | 10 (35.7) | 11 (39.4) | 21 (37.6) |
| Total | 28 (100.0) | 28 (100.0) | 56 (100.0) |
Note: Staphylococcus hemolyticus was the most widely represented species in these 28 cases of CoNS bacteremia (39.3% of blood cultures and 42.8% of stool cultures) followed by uCoNS (35.7% of blood cultures and 39.4% of stool cultures); (*): S. Staphylococcus; **uCoNS: Unidentified Coagulase-Negative Staphylococcus.
Table 3: Distribution of coagulase-negative Staphylococcus strains isolated from blood culture and stool culture.
Antibiotics susceptibility of CoNS strains
One hundred percent of the 28 CoNS strains from blood samples were resistant to penicillin, cefoxitin and kanamycin and 96.4% of isolates were resistant to gentamicin, tobramycin and netilmicin (Table 4). The distribution of resistance patterns of these isolates showed sixteen antimicrobial Resistance (R) patterns a to p and 10 of these strains exhibited R-pattern e (35.7%) and corresponding to isolates 5-11, 13-15.
| Antibiotics tested | S. haemolyticus N= 11 | S. epidermidis N=5 | uCONS N=10 | S. capitis N=1 | S. warneri N=1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | R | S | R | S | |
| Penicillin | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 | 0 |
| Cefoxitin | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 | 0 |
| Kanamycin | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 | 0 |
| Gentamicin | 11 (100) | 0 | 5 (100) | 0 | 8 (80) | 2 (20) | 1 | 0 | 1 | 0 |
| Tobramycin | 11 (100) | 0 | 5 (100) | 0 | 8 (80) | 2 (20) | 1 | 0 | 1 | 0 |
| Netilmicin | 11 (100) | 0 | 4 (80) | 1 (20) | 8 (80) | 2 (20) | 1 | 0 | 1 | 0 |
| Erytromycin | 11 (100) | 0 | 4 (80) | 1 (20) | 6 (60) | 4 (40) | 0 | 1 | 0 | 1 |
| Lincomycin | 1 (9.1) | 10 (90.9) | 2 (40) | 3 (60) | 1 (10) | 9 (90) | 0 | 1 | 0 | 1 |
| Pristinamycin | 0 | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 |
| Rifampin | 10 (90.9) | 1 (9.1) | 4 (80) | 1 (20) | 6 (60) | 4 (40) | 1 | 0 | 1 | 0 |
| Ofloxacin | 11 (100) | 0 | 4 (80) | 1 (20) | 7 (70) | 3 (30) | 0 | 1 | 0 | 1 |
| Vancomycin | 0 | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 |
| Fucidic acid | 0 | 11 (100) | 1 (20) | 4 (80) | 4 (40) | 6 (60) | 0 | 1 | 0 | 1 |
| Trimethoprim sulfamethoxazole | 0 | 11 (100) | 0 | 5 (100) | 0 | 10 (100) | 0 | 1 | 0 | 1 |
| Fosfomycin | 1 (9.1) | 10 (90.9) | 0 | 5 (100) | 3 (30) | 7 (70) | 1 | 0 | 0 | 1 |
| Doxycyclin | 1 (9.1) | 10 (90.9) | 2 (40) | 3 (60) | 1 (10) | 9 (90) | 0 | 1 | 0 | 1 |
Note: Antibiotic results were expressed as susceptible or resistant according to AC-FMS/EUCAST criteria: Inhibition diameters: Penicillin (R<26 mm); Cefoxitin (R<24 mm); Kanamycin (R<14 mm); Gentamicin (S ≥ 22 mm, R<22 mm); Tobramycin (S ≥ 22 mm, R<22 mm); Netilmicin (S ≥ 22 mm, R<22 mm); Erythromycin (S ≥ 21 mm, R<18 mm); Lincomycin (S ≥ 22 mm, R<19 mm); Pristinamycin (S ≥ 21 mm, R<18 mm); Rifampin (S ≥ 26 mm, R<23 mm); Ofloxacin (S ≥ 20 mm, R<20 mm); Vancomycin (S ≤ 2 mg/L); Fusidic acid (S ≥ 24 mm, R<24 mm); Trimethoprim-sulfamethoxazole (S ≥ 17 mm); Fosfomycin (S ≥ 6 mm, R<6 mm); Doxycycline (S ≥ 22 mm; R<19 mm); High resistance: 90.9-100%; Moderate resistance: 70-80%; Low resistance: ≤ 60%; High susceptibility: 90-100%; Moderate susceptibility: 70-80%; Low susceptibility: ≤ 60%; R: Resistant; S: Susceptible; uCoNS: Unidentify Coagulase-Negative Staphylococcus.
Table 4: Antibiotic susceptibility of Coagulase-Negative Staphylococcus (CoNS) strains isolated from blood samples.
One hundred percent of the all 28 CoNS strains isolated from stool (100%) were resistant to penicillin, cefoxitin and kanamycin and 96.4% of the strains were also resistant to gentamicin, tobramycin and netilmicin (Table 5). All of these isolates were grouped into sixteen R-patterns a to p and 11 of these strains displayed R-patterns e (39.2%) and corresponding to isolates 5-11, 13-16 (Table 6).
| Antibiotics tested | S. haemolyticus N=11 | S. epidermidis N=5 | uCONS N=10 | |||
|---|---|---|---|---|---|---|
| R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | |
| Penicillin | 12 (100) | 0 | 5 (100) | 0 | 11 (100) | 0 |
| Cefoxitin | 12 (100) | 0 | 5 (100) | 0 | 11 (100) | 0 |
| Kanamycin | 12 (100) | 0 | 5 (100) | 0 | 11 (100) | 0 |
| Gentamicin | 12 (100) | 0 | 5 (100) | 0 | 10 (90.9) | 1 (9.1) |
| Tobramycin | 12 (100) | 0 | 5 (100) | 0 | 10 (90.9) | 1 (9.1) |
| Netilmicin | 12 (100) | 0 | 5 (100) | 0 | 10 (90.9) | 1 (9.1) |
| Erytromycin | 12 (100) | 0 | 4 (80) | 1 (20) | 8 (72.7) | 3 (27.3) |
| Lincomycin | 0 | 12 (100) | 3 (60) | 2 (40) | 1 (9.1) | 10 (90.9) |
| Pristinamycin | 0 | 12 (100) | 0 | 5 (100) | 0 | 11 (100) |
| Rifampin | 11 (91.6) | 1 (8.4) | 4 (80) | 1 (20) | 6 (54.5) | 5 (45.5) |
| Ofloxacin | 12 (100) | 0 | 3 (60) | 2 (40) | 9 (81.8) | 2 (18.2) |
| Vancomycin | 0 | 12 (100) | 0 | 5 (100) | 0 | 11 (100) |
| Fucidic acid | 0 | 12 (100) | 1 (20) | 4 (80) | 1 (9.1) | 10 (90.9) |
| Trimethoprim sulfamethoxazole | 0 | 12 (100) | 1 (20) | 4 (80) | 0 | 11 (100) |
| Fosfomycin | 1 (8.4) | 11 (81.6) | 0 | 5 (100) | 3 (27.3) | 8 (72.7) |
| Doxycyclin | 2 (16.6) | 10 (83.4) | 0 | 5 (100) | 3 (27.3) | 8 (72.7) |
Note: Inhibition diameters: Refer Table 3; High resistance: 90.9-100%; Moderate resistance: 72.7-80%; Low: ≤ 60%; High susceptibility: 90.9-100%; Moderate susceptibility: 72.2-80%; Low susceptibility: ≤ 45.5%; N: Number.
Table 5: Antibiotic susceptibility of Coagulase-Negative Staphylococcus (CoNS) strains isolated from stool samples.
| Antimicrobial resistance profiles of CoNS strains isolated from blood samples | R patterns | Number of CoNS isolated from blood samples | Antimicrobial resistance profiles of CoNS strains isolated from stool samples | R patterns | Number of CoNS isolated from stool samples |
|---|---|---|---|---|---|
| PR FOXR KR GR TR NR ER OFXR | a | 1 | PR FOXR KR GR TR NR ER OFXR | a | 1 |
| PR FOXR KR GR TR NR ER LR RIFR OFXR FAR | b | 2 | PR FOXR KR GR TR NR ER LR RIFR OFXR FAR | b | 2 |
| PR FOXR KR GR TR NR ER OFXR | c | 3 | PR FOXR KR GR TR NR ER OFXR | c | 3 |
| PR FOXR KR GR TR NR ER LR RIFR OFXR | d | 4, 24 | PR FOXR KR GR TR NR ER LR RIFR OFXR | d | 4 |
| PR FOXR KR GR TR NR ER RIFR OFXR | e | 5-11, 13-15 | PR FOXR KR GR TR NR ER RIFR OFXR | e | 5-11, 13-15, 16 |
| PR FOXR KR GR TR NR ER LR RIFR OFXR FOSR | f | 12 | PR FOXR KR GR TR NR ER LR RIFR OFXR FOSR | f | 12 |
| PR FOXR KR GR TR NR RIFR FOSR | g | 16, 26 | PR FOXR KR GR TR NR ER OFXR DOXR | g | 17, 19 |
| PR FOXR KR GR TR NR ER RIFR OFXR FAR DOXR | h | 17 | PR FOXR KR GR TR NR ER RIFR OFXR DOXR | h | 18, 20 |
| PR FOXR KR GR TR NR ER RIFR OFXR DOXR | i | 18 | PR FOXR KR GR TR NR ER RIFR | i | 21 |
| PR FOXR KR GR TR NR RIFR DOXR | j | 19 | PR FOXR KR GR TR NR OFXR FAR FOSR | j | 22 |
| PR FOXR KR GR TR NR ER RIFR OFXR FOXR DOXR | k | 20 | PR FOXR KR GR TR NR DOXR | k | 23 |
| PR FOXR KR GR TR NR ER OFXR | l | 21 | PR FOXR KR GR TR NR RIFR | l | 24 |
| PR FOXR KR GR TR NR OFXR FAR FOSR | m | 22 | PR FOXR ER OFXR | m | 25 |
| PR FOXR KR ER FAR | n | 23 | PR FOXR KR GR TR NR ER RIFR OFXR FOSR | n | 26 |
| PR FOXR KR OFXR | o | 25 | PR FOXR KR GR TR NR ER LR RIFR SXTR | o | 27 |
| PR FOXR KR GR TR NR RIFR | p | 27, 28 | PR FOXR KR GR TR NR RIFR OFXR | p | 28 |
Note: Sixteen antimicrobial resistance patterns were observed in CoNS spp. isolated from blood sample cultures, 10 of which exhibited R pattern e (35.7%) (isolates 5-11, 13-15); All CoNS spp. isolated from stool sample cultures were classified into 16 antimicrobial resistance patterns, 11 of which exhibited R pattern e (39.2%) (5-11, 13-16); R : Resistance.
Table 6: Antimicrobial Resistance (R) patterns of CoNS spp. isolated from blood sample cultures and stool sample cultures.
The most prevalent resistance patterns observed in the isolates comprised heterogeneous resistance to methicillin, kanamycin, gentamicin, tobramycin, netilmicin and resistance to erythromycin, rifampin and ofloxacin.
The results of the blood and stool cultures are compared based on the resistance pattern
With 28 Staphylococcus isolates, blood culture results were consistent with stool culture results in 53.5% (15/28) of cases and discordant in 46.5% (13/28) of cases. Ten of the fifteen concordant strains displayed R-pattern e, which corresponded to eight S. haemolyticus and two uCoNS, while five strains displayed the R-patterns a, b, c, d and f, which corresponded to three S. epidermidis, one S. haemolyticus and one uCoNS isolates, respectively.
From the 13 discordant cases, the following R-patterns were detected on blood cultures: g (uCoNS and S. capitis isolate); h (uCoNS isolate); j (S. epidermidis isolate); k (S. haemolyticus isolate); l (uCoNS isolate); m (uCoNS isolate); n (uCoNS isolate); d (S. haemolyticus isolate); o (uCoNS isolate) and p (S. warneri and uCoNS isolates).
From the 13 discordant cases, the following R-patterns were detected on stool cultures: e (uCoNS isolate); g (two uCoNS isolates); h (two S. haemolyticus isolates); i (uCoNS isolate); j (uCoNS isolate); k (uCoNS isolate); l (S. epidermidis isolate); m (uCoNS isolate); n (S. haemolyticus isolate); o (S. epidermidis isolate) and p (uCoNS isolate). S. epidermidis (R-pattern i) and S. haemolyticus (R-pattern h) strains were isolated from blood culture and stool culture of patient 18, respectively.
Molecular typing results
Phenotyping results show that 15 preterm infants have BT from the GIT to the circulatory system. The same Staphylococcus spp. were isolated from stool and peripheral blood and they shared the same resistance pattern and they were further genotyped by ERIC-PCR and RAPD-PCR to confirm BT. Fifteen isolates were selected in order to obtain a diverse sample of patients, blood and stool samples and R- patterns. ERIC-PCR and RAPD-PCR were used to compare these 15 selected Staphylococcus strains. Fifteen selected isolates had three different ERIC patterns (A-C) (Figure 2) and three different RAPD patterns (D-F) (Figure 3). ERIC-2 patterns comprised A (S. epidermidis (isolates 1, 2 and 4)); B (S. haemolyticus (isolates 3, 5-11 and 15)) and C (uCoNS (isolates 12-14)). The RAPD patterns were D (uCoNS (isolates 12-14)), E (S. haemolyticus (isolates 3, 5-11 and 15)) and F (S. epidermidis (isolates 1, 2 and 4)). The three R-patterns of S. epidermidis a, b and d, exhibited the AF genotype. Three other uCoNS strains that had resistance patterns e and f showed the CD genotype. In the end, nine S. haemolyticus phenotype e strains showed the BE genotype. In this major epidemic, the BE profile included 60% (9/15) of S. haemolyticus strains isolated in both blood culture and stool culture. The other three types of S. epidermidis and three types of uCoNS that have the AF and CD genotypes, respectively, were thought only happen occasionally. Both units participating in this study had the BE genotype identified. The fusion of ERIC-2 and RAPD results has revealed the existence of three distinct genomic groups namely gg I to III. The strains isolated from blood culture and stool culture in each group were more similar to each other than to the other groups.
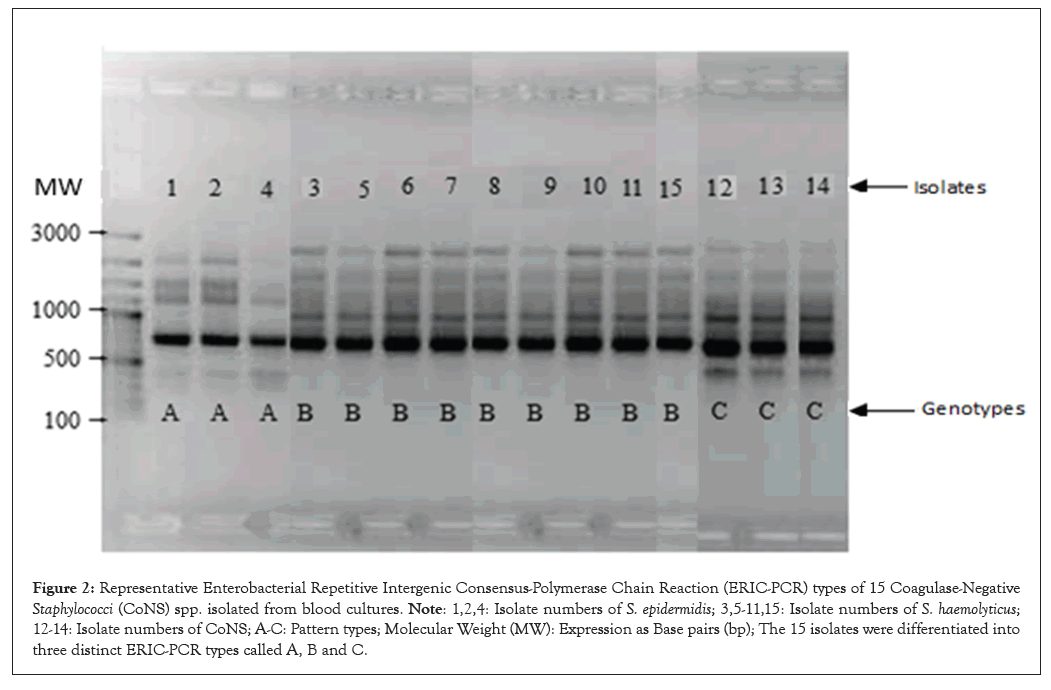
Figure 2: Representative Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR) types of 15 Coagulase-Negative Staphylococci (CoNS) spp. isolated from blood cultures. Note: 1,2,4: Isolate numbers of S. epidermidis; 3,5-11,15: Isolate numbers of S. haemolyticus; 12-14: Isolate numbers of CoNS; A-C: Pattern types; Molecular Weight (MW): Expression as Base pairs (bp); The 15 isolates were differentiated into three distinct ERIC-PCR types called A, B and C.
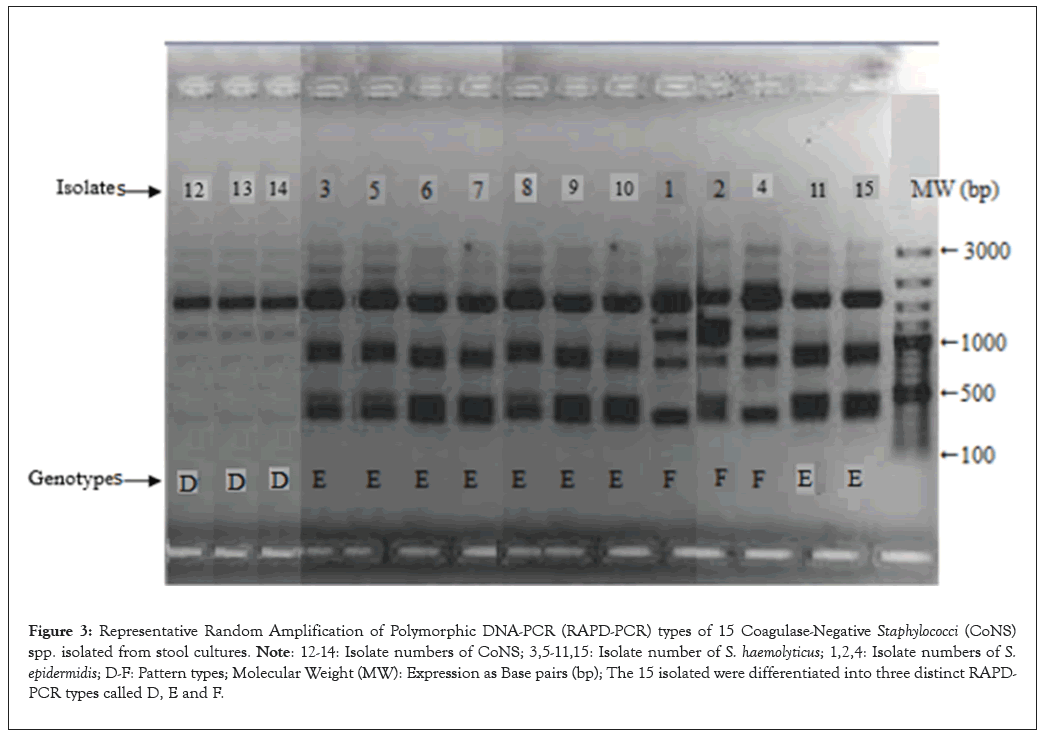
Figure 3: Representative Random Amplification of Polymorphic DNA-PCR (RAPD-PCR) types of 15 Coagulase-Negative Staphylococci (CoNS) spp. isolated from stool cultures. Note: 12-14: Isolate numbers of CoNS; 3,5-11,15: Isolate number of S. haemolyticus; 1,2,4: Isolate numbers of S. epidermidis; D-F: Pattern types; Molecular Weight (MW): Expression as Base pairs (bp); The 15 isolated were differentiated into three distinct RAPD-PCR types called D, E and F.
Bacterial translocation results
In 53.5% of preterm infants, translocation of GIT to the circulatory system was documented. The same Staphylococcus spp. was not found in blood or stool in 46.5% of preterm infants, which strongly suggests that BT is not present in these preterm infants. This means that the intestinal tract wouldn’t be the only or direct source of bacteremia. In patient 18, blood culture was positive for S. epidermidis and stool culture was positive for S. haemolyticus, although a nasopharyngeal sample taken prior to the onset of bacteremia isolated S. epidermidis. Therefore, it is possible that the respiratory tract was the source of bacteremia in this child. However, we didn’t look into this possibility in our study.
Risk factors for the occurrence of BT in preterm infants with CoNS bacteremia
Table 7 shows the difference between documented (group 1) and undocumented (group 2) GIT BT. The wilcoxon-mann-whitney test showed that there were RFs for BT such as BW (p=0.0098); age at onset of infection (p=0.01); TLC/mm³ (p=0.042); lactate/mmol/L (p=0.0002) and the fisher’s exact test showed that intravenous perfusion lipid emulsion (p=0.02), treated patent ductus arteriosus (p=0.03), hemodynamic disorders (p=0.009), history of jaundice (p=0.02) and neonatal antibiotic therapy (p=0.02) were also RFs for BT. None of the other RFs tested were significant.
| Clinical/biological/therapeutic parameters | Documented translocation (n=15) | Undocumented translocation (n=13) | Wilcoxon-mann -whitney test (p-value) | Fisher’s exact test (p-value ) |
|---|---|---|---|---|
| Gestational age (weeks) | ||||
| Mean ± SD | 28.6 ± 3.35 | 29.9 ± 3.40 | P=0.27 | - |
| Median (range) | 28 (25-36) | 29 (25-36) | - | - |
| Birth weight (g) | ||||
| Mean ± SD | 1254.9 ± 644.3 | 1326.2 ± 582.8 | P=0.0098 | - |
| Median (range) | 986 (592-2800) | 1192 (755-2730) | - | - |
| Delivery mode | ||||
| Vaginal | 46.6% (7/15) | 53.8% (7/13) | - | P=1; OR: 0.7878, 95% CI: 0.1323, 4.2017. |
| Cesarean | 53.4% (8/15) | 46.2% (6/13) | - | |
| Gastrointestinal disorders | 80.0% (12/15) | 84.6% (11/13) | - | P=1; OR: 0.7355, 95% CI: 0.0521, 7.7488. |
| Age at onset of infection (days) | ||||
| Mean ± SD | 18.4 ± 13.65 | 1.5 ± 28.61 | P=0.015 | - |
| Median (range) | 13 (7-48) | 9 (3-104) | - | |
| CRP (mg/L) | ||||
| Mean ± SD | 44.8 ± 52.15 | 30.5 ± 31.35 | P=0.59 | - |
| Median (range) | 23 (0-178) | 23.7 (0-89) | - | |
| Leukocytes/mm³ | ||||
| Mean ± SD | 15120 ± 7799.0 | 39061.5 ± 70588 | P=0.042 | - |
| Median (range) | 12000 (1900-34200) | 18200 (5000-271000) | - | |
| Platelets/mm³ | ||||
| Mean ± SD | 142733 ± 8385 | 181000 ± 11705 | P=0.33 | - |
| Median (range) | 136000 (25000-316000) | 185000 (12000-447000) | - | |
| Lactates/mmol/L | ||||
| Mean ± SD | 6.36 ± 1.30 | 4.04 ± 0.90 | P=0.0002 | - |
| Median (range) | 6.8 (3.5-7.9) | 4.0 (2.9-5.9) | - | |
| Lipid emulsion | 12 (80%) | 4 (30.7%) | - | P=0.02; OR: 8.1821, 95% CI: 1.2555, 73.4536. |
| Antenatal corticosteroid | 14 (93.4%) | 11 (84.6%) | - | P=0.58; OR: 2.4627, 95% CI: 0.1141, 160.529. |
| Proton pump inhibitor | 13 (86.6%) | 10 (76.9%) | - | P=1; OR: 0.6605, 95% CI: 0.01, 14.4325. |
| Treated patent ductus arteriosus | 7 (46.6%) | 10 (76.9%) | - | P=0.03; OR: 0.0961, 95% CI: 0.0018, 0.9895. |
| Hemodynamic disorders | 10 (66.6%) | 2 (15.3%) | - | P=0.009; OR: 9.9418, 95% CI: 1.3872, 127.034. |
| History of jaundice | 14 (93.3%) | 7 (53.8%) | - | P=0.02; OR: 10.9341, 5% CI: 1.0273, 587.9893. |
| Antenatal antibiotic therapy | 9 (60%) | 7 (53.8%) | - | P=1; OR: 1.2742, 95% CI: 0.2241, 7.3857. |
| Neonatal antibiotic therapy | 12 (80%) | 4 (30.7%) | - | P=0.02; OR: 8.1821, 95% CI: 1.2555, 73.4236. |
Note: APUH: Amiens Picardy University Hospital; SD: Standard Deviation; CRP: C-Reactive Protein; OR: Odds Ratio; CI: Confidence Intervals; RR: Relative Risk; P: Probability; n: Number.
Table 7: Rates and relative risk factors for digestive bacterial translocation in preterm infants hospitalized in Amiens Picardy University hospital belonging to two groups: Documented translocation and undocumented translocation.
In this study, S.haemolyticus and S.epidermidis were most isolated CoNS spp. with rates of 41.8% and 17.8% respectively. The presence of these pathogens is a major contributor to nosocomial bacteremia and catheter infections in NICUs [29]. Baltimore RS reported that the rate of Nosocomial Infection (NI) in the NICU was 15 to 20% [30]. The literature shows that NIs are common in neonatology, especially in preterm infants who have several RFs such as bacterial colonization, immaturity of the immune system, immaturity of the skin mucous barrier and digestive translocation. Other factors were central catheters, peripheral venous catheters, artificial ventilation using a probe and continuous positive pressure ventilation (nasal or mask) [31].
BT is frequently encountered in patients with critically ill patients [32]. The concept has changed since Berg, et al., [33] first defined as the presence of bacteria in cultured MLN. BT is a normal process that our immune cells control. However, it can become dangerous in cirrhosis when the immune system is not working properly, resulting in increase in its. Moreover, the concept has evolved beyond the translocation of viable bacteria, also considering different bacterial immunologic products without over infection. This concept evolved from evidence that showed a significantly increased systemic pro-inflammatory response in a subgroup of non-infected cirrhotic patients, reaching levels seen in patients with spontaneous bacterial peritonitis. Life-threatening complications in cirrhosis have been described as a result of the translocation of bactDNA [34]. Considering that these patients don’t have any obvious infections, Rodriguez-Laiz, et al., [32] proposed that the presence of bacterial immunogenic products, such as bactDNA, may indirectly reflect certain unfavorable inflammatory conditions that facilitate the development of complications in patients.
The impact of BT on neonates is largely unknown, despite its potential significance in the morbidity and mortality of adults exposed to multiple forms of stress. More and more evidence has shown that normal as well as premature or ill neonates often get BT spontaneously. In this article, we review the recent literature on BT in stress adults and the role of spontaneous BT in the development of sepsis in preterm neonates.
This study identified the main RFs associated with BT as cofounding, including BW (p=0.0098), TLC/mm³ (p=0.042), age at onset of infection (p=0.015), lactates/mmol (p=0.0002), hemodynamic disorders (p=0.009) and history of jaundice (p=0.02). However, there are three independent RFs: intravenous lipid emulsion perfusion (p=0.02), treatment patent ductus arteriosus (p=0.03) and neonatal antibiotic therapy (p=0.02). Doit, et al., [31] found that GA was cofunding RFs and proton pump use was independent RFs, contrary to our study, where these two parameters were not found as RFs. This study also showed that BT in preterm infants with sepsis is responsible for secondary bacteremia and is driven by external factors, such as how long the infants stay in the NICU or how long the infants are fed by an enteral feeding tube. Jezioski, et al., [35] found these results.
In humans, factors independently associated with BT were intestinal obstruction, jaundice, inflammatory bowel disease, malignancies, preoperative Total Parenteral Nutrition (TPN) and emergency surgery according to MacFie J, et al., [36]. These results are different from those of our study because our study population was only a sample of preterm infants, whereas the participants in macfie’ s study with BT were all 71 years, on average.
Lee, et al., [8] used PCR to confirm that BT from the gut to the blood. Compared to preterm infants without sepsis and term infants, VLBW preterm infants with sepsis had lower microbial diversity in the gut and those without invasive infections showed higher microbial diversity in the gut at birth. The strain-specific PCR confirmed the presence of causative pathogens in the gut in 37.5% of the VLBW preterm with sepsis before or at the onset of sepsis and the persistence of colonization for weeks after antibiotic treatment. The authors conclude that, preterm infants with dysbiosis in their digestive tract are at risk for neonatal sepsis and the causing pathogens may originate from the digestive tract and persist to spread horizontally. There is not enough information about how gut dysbiosis leads to sepsis or what key components of a healthy microbiota prevent infections. The same authors, found bacteria in babies who were born prematurely [8]. Nine had blood stream infection and 3 had central nervous system. These bacteria included gram-negative bacilli (25%): K. pneumoniae, E. coli and Serratia liquefaciens; gram-positive cocci (67.7%): S. capitis, S. epidermidis and S. aureus; gram-positive bacilli (8.3%): Bacillus cereus.
There are many factors than can affect the development of gut microbiota in infants, including the mode of delivery, antibiotic exposures, GA and diet [37-42]. The connection between GIT microbiome and neonatal sepsis is still a mystery. Some studies suggested that intestinal imbalances may be the precursor to the onset of neonatal sepsis [43].
In newborns in advance of neonatal sepsis, the gut microbiota was different depending on timing and areas studied. The previous research has shown that a low diversity of the gut microbiota, possibly due to prolonged antibiotic use, may be associated with sepsis. Babies who were antibiotics in the first 2 days of life had more proteobacteria and less actinobacteria, mostly Bifidobacterium and Lactobacillus. Lee, et al., [8] study supports these notions, which demonstrate that preterm infants with invasive infections have lower diversity in microbiota after birth. The microbiota diversity was negatively correlated with the duration of antibiotic use. The prolonged antibiotic uses significantly reduce the beneficial Bifidobacterium and Lactobacillus in the gut. Thus, the development of neonatal sepsis is influenced by gut dysbiosis.
The molecular typing results in this study revealed that the BT from the gut to the circulatory system was observed in 53.5% of cases. This result is in line to that observed by Balzan, et al., [44], (59%) surpassing the results of Rodriguez, et al., [32] (26%) and Moharem, et al., [45] (33%). O’Boyle, et al., [46] found BT in 15.4% of patients who had a laparotomy. Bellot, et al., [34] found BT in 38% of patients with cirrhotic bowel disease.
In this series of 28 CoNS strains, the predominant antibiotic resistance pattern consisted of 10 strains, including 8 S. haemolyticus strains, mainly presenting a major epidemic profile, including the BE genotype, which rapidly spread after emergence in the two units participating in this study. This major epidemic pattern, corresponds to 28.6% (8/28) of the isolated cases. Furthermore, a close correlation was observed between the various genotypes and the associated antibiotic resistance patterns, indicating the emergence of multidrug resistance (resistance to β-lactams, to aminoglycosides and to fluoroquinolones). The increasing use of β-lactams and aminoglycosides contributes to the antibiotic resistance of these strains.
The findings of this study indicate that CoNS spp. which are a component of intestinal microbiota, were the most probable cause CoNS bacteremia in hospitalized preterm infants. The present study demonstrated BT primarily through the comparison of the ERIC-PCR and RAPD-PCR genomic patterns of the identical Staphylococcus spp., which were simultaneously from the blood and stool. Reddy, et al., [47] have compared DNA fingerprints made by amplifying regions DNA with ERIC-PCR and Repetitive Extragenic Palindromic (REP) sequences. Nine of the MLN samples of 98 patients had E. coli. Authors compared the DNA fingerprints of MLN isolates with feces samples from the same patient. All the nine E. coli strains isolated from MLNs were found to possess identical DNA fingerprints. This study supports the hypothesis that BT originates in intestine. A useful technique to establish relatedness between stool and blood isolates from children have been shown to be PFGE [48]. The relationship between microbiologic culture and sequencing analysis has been demonstrated by Rodriguez-Laiz, et al., [32]. These authors identified CoNS spp. in 35.8% of cases using both microbiologic culture and molecular tools, 42.8% were identified using molecular tools only and 10.7% with microbiological culture only [32]. It is hard to show a connection between BT and bacteremia. The clinical consequences of this bacteremia only appear to be heightened by massive BT, when the body’s ability to eliminate the pathogen is surpassed, when the pathogen is especially aggressive or the host’s immune system is disrupted [45,48]. Under these conditions, micro-organisms from the GIT may cause localized or generalized infections. The latest advances in molecular biology allow for noninvasive investigation BT within humans [35,49].
Limitations
Our study has its limitations, such as the small sample size, which limits interference with regard to causality. This study was conducted on 28 preterm infants with at least bacteremia and a positive CoNS stool culture. This inclusion criterion is extremely restrictive as this study solely focused on BT from intestinal origin. This study didn’t examine other areas, such as the respiratory or the skin. Staphylococcus spp. are the predominant microbe present in the microbiota of preterm infants, thus it can be inferred that all preterm infants are carriers of detectable CoNS or not in standard culture. This sample of patients can be considered a random sample of preterm newborns with CoNS bacteremia.
The GIT is a multifaceted organ involved in numerous physiological functions. The immune system and microbiota in the GIT are crucial for neonatal sepsis. The pathogenesis of sepsis involves BT, host and environmental factors. This study showed that preterm infants with gut dysbiosis are more likely to get neonatal sepsis. It is possible that the causative CoNS is bacteria that previously colonize in the gut or acquired in the environment. The RF for gut dysbiosis in preterm infants is prolonged exposure to antibiotic. This study shows that BT starts in the GIT. Most translocating CoNS are normal commensal microflora and do not belong to any known pathotype. Our result suggests that BT is more dependent on the gut epithelium than the virulence of the resident enteric bacteria. ERIC-PCR and RAPD-PCR showed that BT was present in 15 patients who fit certain characteristics. The CoNS isolated from blood and stool showed a high degree of phenotypic and genotypic similarity in 53.5% of cases. The stools of this group of patients with CoNS bacteremia were a definitive source of CoNS. BT could not formally be excluded from 46.5% of discordant cases with or without bacteremia obtained from blood cultures and stool cultures, because there were other non-intestinal sources that were not analyzed in this study.
The research protocol used in studies involving human subjects were in line with the guidelines laid down in the Declaration of Helsinki and approved by the local Person Protection Committee (PPC) (Ethical Committee), Ile de France 3, ID-RCB; AO3540-53, opinion favorable, ref. DS/LG/2018-039. This study is retrospective and non-interventional that does not involve the human person and is included exclusively clinical and laboratory data concerning preterm infants, for this reason this study did not ask for consent from the legal guardians of the minors who were included. The study participants were all breastfeeding mothers who were considered to be autonomous and independent and able to decide whether or not to let medical data of their baby take part in the study.
All authors have no conflicts of interest to disclose.
The authors don’t have any other interests that are related to this article’s content.
The authors haven’t disclosed any financial relationships related to this article.
Leke A, Amar G, Morin G, Goudjil S, Kongolo G and Biendo M participated in meetings and discussions that led to this manuscript. They contributed to the study conception, design and drafting of the manuscript. All authors helped to get, analyzed and interpreted data. They also helped to edit and make final manuscript. The final manuscript was reviewed and approved by all authors.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Leke A, Kongolo G, Frere-Moysan M, Ghostine G, Chazal C, Biendo M (2024) The Value of Molecular Typing in the Assessment of Bacterial Translocation During Coagulase-Negative Staphylococcal Bacteremia in Preterm Infants. Adv Pediatr Res. 11:080.
Received: 14-May-2024, Manuscript No. LDAPR-24-30127; Editor assigned: 16-May-2024, Pre QC No. LDAPR-24-30127 (PQ); Reviewed: 31-May-2024, QC No. LDAPR-24-30127; Revised: 07-Jun-2024, Manuscript No. LDAPR-24-30127 (R); Published: 14-Jun-2024 , DOI: 10.35248/2385-4529.24.11.080
Copyright: © 2024 Leke A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.