Journal of Nanomedicine & Biotherapeutic Discovery
Open Access
ISSN: 2155-983X
ISSN: 2155-983X
Research Article - (2015) Volume 5, Issue 3
Keywords: Benzimidazole; Quantitative structure activity relationship (QSAR); Antimicrobial activity; Multiple linear regressions
Infectious diseases caused by bacteria, fungi, viruses and parasites are still a major threat to public health, despite tremendous progress in medicinal chemistry. The impact is more acute in developing countries due to nonavailability of desired medicines and emergence of widespread drug resistance [1]. Despite this increasing problem of antibiotic resistance, the number of different antibiotics available is dwindling and there are only a handful of new antibiotics in the drug development pipeline. Therefore, there is an urgent need for new antibacterial drugs preferably with new modes of action to potentially avoid cross-resistance [2,3]. Specifically, multi-drug-resistant Grampositive bacteria including methicillin-resistant Staphylococcus aureus (MRSA), Staphylococcus epidermidis (MRSE) and vancomycin resistant enterococci (VRE) are of major concern [4]. The bacterial resistance to antibiotics is a major health problem and resulted in emergence of an endless area for study and debate. Fluconazole is a well known firstline antifungal drug recommended by World Health Organization and is prevalently used to treat fungal infections by Candida albicans and Cryptococcus neoformans with an exceptional therapeutic record including good activity and favourable pharmacokinetic characteristics [5]. Some Fluconazole analogues such as Ravuconazole, Albaconazole and Isavuconazole have been successfully developed as clinical drugs for the treatment of fungal infections [6,7]. QSAR methods have been widely applied for drug discovery, lead optimization, risk assessment, toxicity prediction and regulatory decisions. The success of QSAR studies mainly depends on whether or not the molecular descriptors chosen are appropriate to explain the biological activity. Obtaining a good quality QSAR model depends on many factors, such as the quality of biological data, the choice of descriptors, and statistical methods. QSAR methods are based on statistically determined linear or nonlinear functional forms that relate the activity of interest with descriptors [8,9].
Once a reliable QSAR model is established, we can predict the activities of molecules, and know which structural features play an important role in the biological process. In an attempt to identify the physicochemical and structural features of benzimidazole type of fluconazole analogues adducts required or responsible for potent antimicrobial activity, two-dimensional quantitative structure–activity relationship (2D-QSAR) studies were undertaken in the present work.
Data sets and biological activity
In the present study, a series of novel series of benzimidazole type of Fluconazole analogues having antibacterial and antifungal activity from published result [10] was taken. The biological activity values represented as minimum inhibitory concentrations (MIC) were first converted to –log MIC (pIC50) in micromolar (Table 1) and were used as the dependent variable for the QSAR model. The QSAR models were generated using a training set of twenty seven molecules and remaining seven compounds as a test set (Table 1 marked with asterisk) for validating the quality of the models. The structures of the compounds in the training and test sets are shown in (Table 1).
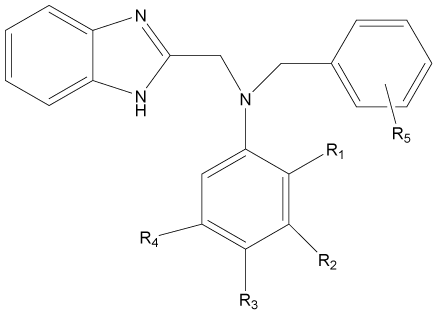 |
|||||||||
| S. No | R1 | R2 | R3 | R4 | R5 | MICa | MICb | MICc | MICd |
| 1 | F | H | H | H | H | 4.798 | 4.795 | 4.494 | 4.494 |
| 2 | H | CF3 | H | H | H | 4.795 | 4.494 | 4.494 | 4.494 |
| 3* | H | H | F | H | H | 4.494 | 4.193 | 4.795 | 4.795 |
| 4 | H | H | Cl | H | H | 4.193 | 4.494 | 4.494 | 4.795 |
| 5 | H | H | I | H | H | 4.193 | 3.892 | 4.795 | 4.795 |
| 6 | H | H | CH3 | H | H | 4.494 | 3.290 | 4.795 | 4.494 |
| 7 | F | H | F | H | H | 4.795 | 4.795 | 4.795 | 4.795 |
| 8* | Cl | H | Cl | H | H | 4.193 | 4.795 | 4.494 | 4.494 |
| 9 | H | CF3 | H | CF3 | H | 5.698 | 5.096 | 5.698 | 5.698 |
| 10 | 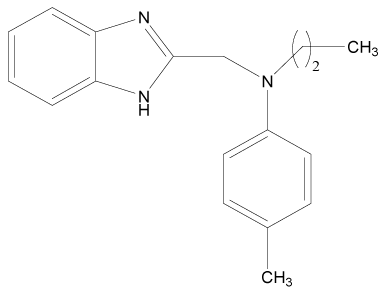 |
H | H | H | H | 3.892 | 3.892 | 3.892 | 3.892 |
| 11 | 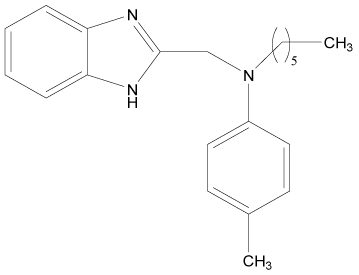 |
H | H | H | H | 3.892 | 4.795 | 3.591 | 4.795 |
| 12* | 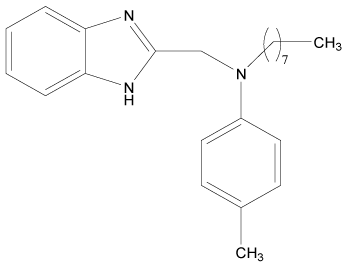 |
H | H | H | H | 4.494 | 3.892 | 3.892 | 4.795 |
| 13 | 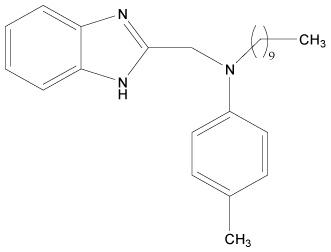 |
H | H | H | H | 3.892 | 3.892 | 3.892 | 4.795 |
| 14 | 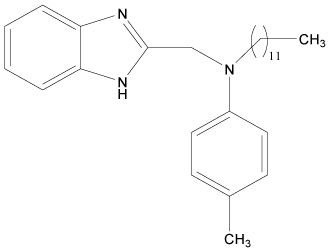 |
H | H | H | H | 4.193 | 4.795 | 3.892 | 4.795 |
| 15 | 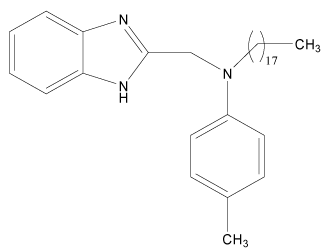 |
H | H | H | H | 3.892 | 3.591 | 3.591 | 4.494 |
| 16 | F | H | H | H | 3, 4-2 Cl | 4.193 | 3.892 | 4.494 | 4.494 |
| 17* | H | CF3 | H | H | 2, 4-2 Cl | 4.494 | 4.494 | 4.795 | 4.795 |
| 18 | H | H | F | H | 2, 4-2 Cl | 4.494 | 4.795 | 3.892 | 4.494 |
| 19 | H | H | CH3 | H | 2-Cl | 4.193 | 3.892 | 4.494 | 4.494 |
| 20 | H | H | CH3 | H | 3-Cl | 4.494 | 3.892 | 4.494 | 4.494 |
| 21* | H | H | CH3 | H | 4-Cl | 4.494 | 3.892 | 4.494 | 4.795 |
| 22 | H | H | CH3 | H | 4-NO2 | 4.494 | 3.892 | 4.795 | 3.892 |
| 23 | H | H | CH3 | H | 2, 4-2F | 4.795 | 4.795 | 3.892 | 4.494 |
| 24 | H | H | CH3 | H | 2, 4-2 Cl | 4.193 | 3.892 | 4.494 | 4.795 |
| 25 | H | H | CH3 | H | 3, 4-2 Cl | 4.494 | 4.795 | 4.795 | 3.892 |
| 26 | F | H | F | H | 2, 4-2 Cl | 4.193 | 4.795 | 4.494 | 3.892 |
| 27 | Cl | H | Cl | H | 2, 4-2F | 4.193 | 3.892 | 4.795 | 3.892 |
| 28* | H | CF3 | H | CF3 | 2, 4-2F | 5.096 | 5.096 | 4.795 | 4.795 |
| 29 | H | CF3 | H | CF3 | 2, 4-2 Cl | 4.193 | 3.892 | 4.795 | 4.795 |
| 30 | 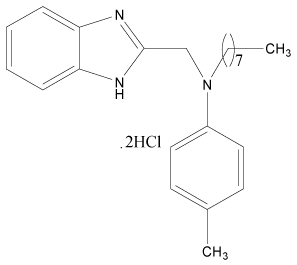 |
H | H | H | H | 4.193 | 4.795 | 3.892 | 4.795 |
| 31 | 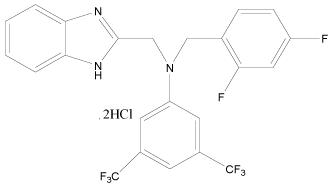 |
H | H | H | H | 5.096 | 5.096 | 4.494 | 5.096 |
| 32* | 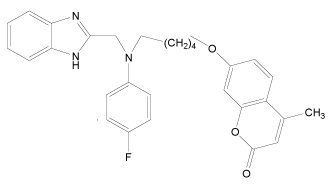 |
H | H | H | H | 4.494 | 3.892 | 3.892 | 4.795 |
| 33 | 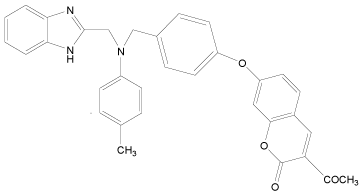 |
H | H | H | H | 3.892 | 3.892 | 3.892 | 4.795 |
| 34 | 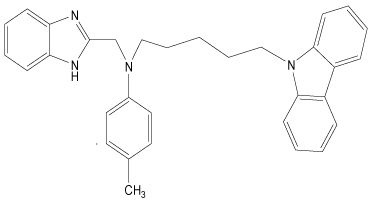 |
H | H | H | H | 4.193 | 3.892 | 3.892 | 4.494 |
Table 1: Structures and antimicrobial activities of benzimidazole type of Fluconazole analogues.
Calculations of 2D-QSAR descriptors
2D-QSAR studies were carried out on a Windows XP workstation using the molecular modeling software package VLife Molecular Design Suite (VLife MDS) version 3.5 [11]. Compounds were sketched using the 2D draw application and converted to 3D structures. Energy minimization and geometry optimization were conducted using Merck molecular force field (MMFF) and atomic charges, maximum number of cycles were 1000, convergence criteria (RMS gradient) was 0.01 and medium’s dielectric constant of 1 by batch energy minimization method [12].
A set of quantum mechanical descriptors: Energy for various 2D descriptors Individual, Chi, Path Count, Element Count, Estate Numbers, and Polar Surface Area and Alignment independent and these calculated descriptors were considered as independent variables in this study. The preprocessing of the independent variables (i.e., 2D descriptors) was done by removing invariable (constant column), which resulted in total 210 descriptors to be used for QSAR analysis.
In this study, three 2D-QSAR models were obtained which are discussed below
pIC50 = 1.0542 (±0.3275) SsOHcount -0.8939 (±0.2616) HUMO energy +0.4216 (±0.0329) T_N_F_1
N= 34 r2 = 0.7927, q2 = 0.7242, F test = 52.5423, pred_r2 = 0.7531, pred_r2se = 0.216
The significance of the quantitative correlation between the descriptor and the biological activity are expressed as correlation coefficient (r2 =0.7927), cross validated correlation coefficient (q2= 0.7242). The alignment independent descriptor T_N_F_1 indicate the presence of fluorine atom (i.e. –F or -CF3) at R1, R2, R3, R4 and R5 position on ring to be detrimental for the activity. Model shows the positive contribution of SsOHcount shows that the activity was increased with the presence of hydroxy groups in moiety. The correlation matrix is shown in (Table 2). The observed and predicted activity values of the training and test set compounds selected for the model are given in (Table 3). The activity contribution chart for 2D-QSAR model is shown in Figure 1a and plots of observed vs. predicted values of pIC50 are shown in Figure 1b.
| Parameter | SsOHcount | HUMO energy | T_N_F_1 |
| SsOHcount | 1.0000 | ||
| HUMO energy | -0.3643 | 1.0000 | |
| T_N_F_1 | 0.4187 | 0.5597 | 1.0000 |
Table 2: Correlation Matrix for 2D best Model 1.
| Com | S. aureus Model-1 | M. luteus Model-2 | E. coli-model-3 | C. albicans-model-4 | ||||
|---|---|---|---|---|---|---|---|---|
| Pred. | Res. | Pred. | Res. | Pred. | Res. | Pred. | Res. | |
| 1 | 4.326 | 0.472 | 4.741 | 0.054 | 4.461 | 0.033 | 4.513 | -0.019 |
| 2 | 4. 852 | -0.057 | 4.514 | -0.02 | 4.474 | 0.02 | 4.466 | 0.028 |
| 3 | 4.374 | 0.12 | 4.183 | 0.01 | 4.813 | -0.018 | 4.813 | -0.018 |
| 4 | 4.087 | 0.106 | 4. 462 | 0.032 | 4.487 | 0.007 | 4.687 | 0.108 |
| 5 | 4.329 | -0.136 | 3.956 | -0.064 | 4.831 | -0.036 | 4.771 | 0.024 |
| 6 | 4.299 | 0.195 | 3.216 | 0.074 | 4.708 | 0.087 | 4.342 | 0.152 |
| 7 | 4.582 | 0.213 | 4.769 | 0.026 | 4.921 | -0.126 | 4.828 | -0.033 |
| 8 | 4.046 | 0.147 | 4.706 | 0.089 | 4.528 | -0.034 | 4.467 | 0.027 |
| 9 | 5.825 | -0.127 | 5.149 | -0.053 | 5.645 | 0.053 | 5.645 | 0.053 |
| 10 | 3.625 | 0.267 | 3.791 | 0.101 | 3.759 | 0.133 | 3.936 | -0.044 |
| 11 | 3.941 | -0.049 | 4.626 | 0.169 | 3.684 | -0.093 | 4.808 | -0.013 |
| 12 | 4.174 | 0.32 | 3.886 | 0.006 | 3.929 | -0.037 | 4.824 | -0.029 |
| 13 | 3.661 | 0.231 | 3.933 | -0.041 | 3.646 | 0.246 | 4.737 | 0.058 |
| 14 | 4.083 | 0.11 | 4.915 | -0.12 | 3.788 | 0.104 | 4.723 | 0.072 |
| 15 | 3.738 | 0.154 | 3.648 | -0.057 | 3.589 | 0.002 | 4.381 | 0.113 |
| 16 | 4.365 | -0.172 | 3.691 | 0.201 | 4.364 | 0.13 | 4.469 | 0.025 |
| 17 | 4.364 | 0.13 | 4.431 | 0.063 | 4.717 | 0.078 | 4.837 | -0.042 |
| 18 | 4.593 | -0.099 | 4.631 | 0.164 | 3.911 | -0.019 | 4.763 | -0.269 |
| 19 | 4.013 | 0.18 | 3.712 | 0.18 | 4.562 | -0.068 | 4.391 | 0.103 |
| 20 | 4.249 | 0.245 | 3.926 | -0.034 | 4.359 | 0.135 | 4.374 | 0.12 |
| 21 | 4.383 | 0.111 | 3.819 | 0.073 | 4.536 | -0.042 | 4.831 | -0.036 |
| 22 | 4.413 | 0.081 | 3.708 | 0.184 | 4.872 | -0.077 | 3.866 | 0.026 |
| 23 | 4.681 | 0.114 | 4.816 | -0.021 | 3.767 | 0.125 | 4.478 | 0.016 |
| 24 | 4.333 | -0.14 | 3.967 | -0.075 | 4.379 | 0.115 | 4.683 | 0.112 |
| 25 | 4.438 | 0.056 | 4.671 | 0.124 | 4.882 | -0.087 | 3.922 | -0.03 |
| 26 | 4.296 | -0.103 | 4.914 | -0.119 | 4.339 | 0.155 | 3.851 | 0.041 |
| 27 | 4.037 | 0.156 | 3.799 | 0.093 | 4.813 | -0.018 | 3.830 | 0.062 |
| 28 | 5.333 | -0.237 | 5.108 | -0.012 | 4.731 | 0.064 | 4.787 | 0.008 |
| 29 | 4.358 | -0.165 | 3.935 | -0.043 | 4.769 | 0.026 | 4.752 | 0.043 |
| 30 | 3. 992 | 0.201 | 4.739 | 0.056 | 3.862 | 0.03 | 4.806 | -0.011 |
| 31 | 4.964 | 0.132 | 5.018 | 0.078 | 4.516 | -0.022 | 5.186 | -0.09 |
| 32 | 4.281 | 0.213 | 3.909 | -0.017 | 3.824 | 0.068 | 4.653 | 0.142 |
| 33 | 3. 658 | 0.234 | 3.834 | 0.058 | 3. 873 | 0.019 | 4.769 | 0.026 |
| 34 | 4.362 | -0.169 | 3.912 | -0.02 | 3.916 | -0.024 | 4.353 | 0.141 |
Table 3: Predicted activities according to 2D QSAR models results of benzimidazole type of Fluconazole analogues.
pIC50 = 0.3839(± 0.0613) T_2_Cl_1+ +0.1525(± 0.0216) SssOEindex
N=34, r2=0.7416, q2 = 0.6821, F test = 32.5218, pred_r2 = 0.7136
pIC50=0.6531(± 0.1348) SsCH3E-index +0.5821(± 0.065) T_Cl_ Cl_1
N = 34, r2 = 0.7542, q2 = 0.6734, F test = 21.5482, pred_r2 = 0.6642
pIC50=-0.8753(± 0.1641) SddsN (nitro) count +0.1764(± 0.0863) H-Donor Count
N = 34, r2 = 0.7328, q2 = 0.6894, F test = 23.5847, pred_r2 = 0.6222
The model reveals that the descriptor SssOE-index which is electro topological state indices for number of oxygen atom connected with two single bonds showed positive effect indicated that the antimicrobial activity was increased with the presence of methoxy groups of R1 and R3 may lead to an increase in the activity. T_2_Cl_1 indicates that the antimicrobial activity was increase with the presence of chlorine moieties in on the ring will lead to an increase in activity.
The descriptor T_Cl_Cl_1 in a molecule position on ring to be detrimental for the activity. The descriptor SsCH3E-index is topological state indices for number of eCH3 group connected with one single bond. The descriptor signifies the importance of methyl group at R3 positions (compound 19, 20, 21, 22, 23, 24 and 25) has more potent antimicrobial activity.
The descriptor is H-donor count calculates the number of hydrogen atoms in a compound and suggests that increase in H-donor count of moiety R5 may lead to an increase in the activity. The descriptor is SddsN (nitro) count suggests the total number of nitro group connected with one single and two double bonds R5 position will lead to improved activity.
The purpose of the present work was to determine the biological activity of different scaffolds of benzimidazole types of Fluconazole derivatives acting as antimicrobial activity by creating a robust QSAR model. The QSAR results revealed that the presence of methoxy, hydroxy, chloro or fluoro substituents would increase the antimicrobial activity and presence of electron withdrawing groups at R1-R5 position of benzimidazole ring would increase the activity. These models are useful because they rationalize a large number of experimental observations, and allow saving of both time and money in the drug design process.
The author wishes to express gratitude to V-life Science Technologies Pvt. Ltd for providing the trail version software for the study.
The authors declare no conflict of interest.