Journal of Medical Diagnostic Methods
Open Access
ISSN: 2168-9784
ISSN: 2168-9784
Research Article - (2021)Volume 10, Issue 3
Background: HCM is one of the leading causes of sudden cardiac death in adults, this disease is inherited in an autosomal dominant manner, however, the genetic etiology of the disease is not fully explained and studies about hereditary characteristics in family trees are still underway.
Method: Ten HCM patients and 31 of their relatives were recruited for the research. Targeted sequencing for 4 HCM related-genes including MYH7, MYBPC3, TNNT2 and TNNI3 uses targeted Next-Generation Sequencing (NGS). Demographic, clinical, electrocardiography and echocardiography characteristics were also characterized.
Results: Among ten HCM patients, five patients were identified with the HCM pathogenic variants in MYH7 (3 patients), MYBPC3 (1 patient), and TNNT2 (1 patient) genes. Eleven out of thirty-one relatives from these five genotype-positive patients carried the same pathogenic variants. Interestingly, we found the novel c.822-2 A>G variant in the VUS splicing receptor zone of the TNNT2 gene responsible for HCM disease in a family with 7 subjects were genotype-positive, and 3 others suffered from sudden cardiac death.
Conclusions: This case series highlighted the importance of genetic testing for clinically confirmed HCM patients and family members. The genetic information can either be used as a molecular marker to complement clinical presentation in the diagnosis of HCM or a prognosis tool for patients and their family members.
Hypertrophic cardiomyopathy; Vietnamese; MYH7; MYBPC3; TNNT2; TNNI3; Genetic testing; Novel pathogenic variant
HCM: Hypertrophic Cardiomyopathy; LV: Left Ventricular; ECG: Electrocardiography; ESC: European Society of Cardiology; SCD: Sudden Cardiac Death
HCM is recognized as the most common monogenic cardiovascular disease with the prevalence of 1:500 in young adults, it is also one of the leading causes of sudden cardiac death in young adults [1,2]. Hypertrophic Cardiomyopathy (HCM) is defined as the thickening of the myocardial wall in any segments of the Left Ventricular (LV) in the absence of any other causes [3]. HCM is inherited as an autosomal dominant genetic trait that could be transmitted to offspring with a proportion up to 50%. Recent studies have indicated that 60% HCM patients could be identified with the sarcomere disease-causing variant [4,5].
However, the genetic etiology of the disease is not fully explained and studies about the genotype-phenotype correlation are still underway. Up until recently, 11 genes have been identified and extensively studied for its pathogenicity in HCM. Among them, pathogenic variants in the ß-myosin heavy chain (MYH7) and cardiac myosin-binding protein C (MYBPC3) genes are the most commonly associated with HCM, followed by Cardiac Troponin T (TNNT2) and Cardiac Troponin I (TNNI3) genes [6,7].
Recent existing data have been mostly obtained in the Caucasian population while there is a dearth of studies conducted in the Vietnamese population, particularly the population in the North of Vietnam. The study of Thu et al. in 2019, focused on 23 disease related genes has demonstrated a prevalence of HCM-related gene variants in Vietnamese people [8]. However, the hereditary characteristics in families as well as the clinical characteristics of probands and their relatives have not yet been clarified. Therefore, we carried out this study on a series of patients and family members, with an extended Next Generation Sequencing panel covering the 4 most prevalent HCM-related genes (MYH7, MYBPC3, TNNT2, and TNNI3). We also discuss further the extend of penetrance and phenotype-genotype correlation of the variants found in our study and perform a literature review to assess differences across populations.
This study was conducted at Vietnam National Heart Institute, Bach Mai hospital, Hanoi, Vietnam from July 2018 to August 2019. Ten patients diagnosed clinically with HCM were recruited together with 31 family members. The study protocol was approved by the Ethics Committee of Hanoi Medical University (Hanoi Medical University Institutional Review Board, Hanoi, Vietnam, reference number: IRB00003121). The study abides by the Declaration of Helsinki in regards to the study involving human subjects. Informed consents were obtained from all participants.
Clinical assessment
Subjects were clinically diagnosed with HCM by experienced cardiologists and echo-cardiologists in VNHI. The diagnosis criteria were based on ESC guideline 2014-the wall thickness of any LV myocardial segments ≥ 15 mm and no other causes explaining the hypertrophy. For children, the diagnosis of HCM was based on an LV wall thickness more than two standard deviations greater than the predicted mean (z-score>2) [9,10]. Other clinical data includes subjects’ symptoms, medical history, and recorded family history. All subjects had a 12-lead resting electrocardiogram performed by Nihon Koden Model 1250A Cardiofax S. The presence of Atrial Fibrillation (AF), LV hypertrophy, left or right bundle-brand block, left atrial and/or ventricular enlargement, abnormal Q waves and non-specific ST-T segments were recorded. Patients with suspected paroxysmal arrhythmias (Paroxysmal Atrial Fibrillation (PAF), ventricular tachycardia, Premature Ventricular Contractions (PVC), or Block atrio-ventricular) were confirmed with ambulatory electrocardiogram monitored for 24 hours. Echocardiography evaluations for patients and relatives were performed using GE Vivid E9 Ultrasound System and EchoPAC Clinical Workstation Software version V202. All the index including measurements of LV dimensions and wall thickness (septal, posterior, and maximal left ventricular thickness), LV ejection fraction, Right ventricular hypertrophy and Right ventricular dynamic obstruction, LV diastolic function, Left atrium volume index, pulmonary artery systolic pressure, dynamic obstruction at rest combine with Valsalva maneuver, mitral valve and papillary muscle evaluation were evaluated according to recommendations of American Society of Echocardiography in 2011 [11].
Genetic analysis
Upon enrollment, blood samples were obtained from each subject and collected into sterile EDTA tubes for genotyping and laboratory analysis. Genomic DNA was isolated from 2 ml peripheral blood sample obtained from each subject using the Wizard® Genomic DNA Purification Kit (Promega, Madison, USA). DNA was stored at -80°C until the time of analysis. The samples were sent for Next-Generation Sequencing (NGS) of a small panel of 4 genes including MYH7, MYBPC3, TNNT2, and TNNI3. Sanger’s sequencing was the method of choice for verification of identified variants and for testing of family members (Primers’ sequence for Sanger’s sequencing is available upon request)
Variant analysis
Genetics variants found in this study were evaluated for their pathogenicity based on the guideline of the American College of Medical Genetics. Appropriate in silico tools and public domain library (ClinVar, HGMD, LOVD) were utilized to confirm whether if a variant were reported previously. Pymol was utilized to visualize the affected protein as well as the location of the pathogenic variants.
Clinical characteristics, ECG, and echocardiography findings of 16 subjects having genetic positive results were shown in Table 2. We identified four reported pathogenic variants and a new variant of TNNT2 in 5 families. The age of diagnosis for probands with confirmed pathogenic variants were 29 ± 14.7 for 5 patients with HCM-related pathogenic variants and 64.8 ± 4.5 for the group without any suspected pathogenic variant, the difference was significant. Other characteristics of the two group patients were shown in Table 1.
| Proband clinical characteristic | Confirmed pathogenic variant (N=5) | No genetic findings (N=5) | p |
|---|---|---|---|
| Gender, male/female Age of diagnosis, years | 2 males/3 females 29 ± 14.7 | 3 males/2 females 64.8 ± 4.5 | >0.05 0.02 |
| Syncope/Presyncope | 3 | 0 | >0.05 |
| Cardiac symptoms (NYHA = 2/ CCS = 2) | 4 | 4 | >0.05 |
| Family history of SCD | 3 | 1 | >0.05 |
| Family history of HCM | 5 | 3 | >0.05 |
| Maximal LVWT, mm | 17.9 ± 10 | 17.1 ± 1.9 | >0.05 |
| Severe hypertrophic ³ 20 mm | 4 | 3 | >0.05 |
| LVOT pressure, mmHg | 18.2 ± 12.5 | 55.6 ± 48.0 | 0.08 |
| LVOT ³ 30 mmHg | 4 | 4 | >0.05 |
| LVOT ³ 50 mmHg | 4 | 2 | >0.05 |
| SAM | 4 | 4 | >0.05 |
| Heart failure, LVEF < 40% | 1 | 0 | >0.05 |
| ECG abnormalities | 5 | 4 | >0.05 |
| Treatment | |||
| Medications only | 1 | 3 | |
| ASA/Septal myectomy | 4 | 2 | >0.05 |
| ICD | 1 | 0 | |
NYHA, New York Heart Association; CCS, Canadian Cardiovascular Society; SCD, Sudden cardiac death; HCM, Hypertrophic cardiomyopathy; LVWT, Left ventricular wall thickness; LVOT, Left ventricular outflow tract; SAM, Systolic anterior motion; LVEF, Left ventricular ejection fraction; ECG, Echocardiography; ASA, Alcohol septal ablation; ICD, Implantable Cardioverter-Defibrillator.
Table 1: Clinical characteristics of hypertrophic cardiomyopathy patient’s cohort.
Results of genetic analysis
By fully sequenced 4 common genes related to HCM (MYH7, MYBPC3, TNNT2, and TNNI3), we identified genetic variants in 5 families that can be attributed to hypertrophic cardiomyopathy. The variants were p.T1377M, p.R783H, p.R1114H on MYH7; p.D770N on MYBCP3 and c.822-2A>G on TNNT2. Reconfirmation using Sanger’s sequencing was performed on the proband as well as the family’s members to identify all 5 variants found were in the heterozygous state and exist in at least 2 people in each family. The variants found are outlined in Table 2, the primer for Sanger’s sequencing is available from the authors on reasonable request. Four out of five variants have been identified in patients with HCM (p.T1377M, p.R783H, p.R1114H on MYH7; p.D770N on MYBCP3) and one newly identified variant c.822-2A>G on TNNT2. However, we still perform assessments based on ACMG (American College of Medical Genetics) guideline to classify variants’ pathogenicity. In which the four reported variants we assess based on ACMG criteria match with previous pathogenicity reports on databases such as Clinvar, HGMD, or LOVD. The full criteria and classification are included in the supplementary information (Supp. Figure 2).
| Mutation | Relation-ship | Age | Sex | Symptoms | Cardiac Phenotype | ECG | LVWT (mm) | EF (%) | LVOT, mmHg | Management | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | p.T1377M MYH7 | Proband | 62 | F | Dyspnea, syncope | Septal hypokinesis (after ASA) | AF, LBBB | 20 | 58 | 60 | BBs and ASA |
| Son | 40 | M | DOE, chest discomfort | Sigmoid septum with no obstruction | NSR, NSST-T | 18 | 73 | 5 | BBs | ||
| Grandson | 11 | M | None | Normal | NSR | 8 | 82 | 9 | None | ||
| 2 | p.R782H MYH7 | Proband | 39 | M | Dyspnea, oedema, syncope | Septal hypokinesis (after ASA) | BAV III PMR VT in Holter 24h | 22 | 37 | 55 | BBs, ASA and ICD implantation |
| Brother | 27 | M | DOE, chest discomfort | Septum and apex hypertrophy, no obstruction | NSR, NSST-T | 19 | 61 | 8.5 | BBs | ||
| 3 | c.822-2A>G TNNT2* | Proband | 25 | F | DOE, chest pain | Both ventricular hypertrophy with LVOT | RBBB | 24 | 58 | 110 | Medical treatment for heart failure, BBs and ASA |
| Father | 54 | F | DOE, syncope | RV hypertrophy | NSR RVH | 15 | 65 | None | Medical treatment for heart failure | ||
| Aunt | 52 | M | DOE | RV hypertophy | BAV III | 14 | 68 | None | Pace-maker implantation, BBs | ||
| Aunt | 37 | F | DOE | Septal and RV hypertrophy | NSR NSST-T | 13.4 | 66 | 10 | BBs | ||
| Uncle | 41 | M | None | Normal | NSR, NSST-T | 7 | 80 | None | None | ||
| Cousin | 3 | F | None | Normal | Normal | 5 | 71 | None | None | ||
| Cousin | 1 | M | None | Normal | Normal | 6 | 65 | None | None | ||
| 4 | p.D770N MYBPC3 | Proband | 18 | F | DOE, palpitations | Sigmoid septum, no obstruction | NSST-T PAF in Holter 24h | 24 | 74 | 12 | BBs, Acenocoumarol |
| Father | 52 | M | None | Normal | NSR RBBB | 10 | 69 | None | BBs | ||
| 5 | p.R1114H MYH7 | Proband | 30 | M | Dyspnea, chest pain, Syncope | Symmetric hypertrophy | NSTT-T | 31 | 80 | 50 | BBs and ASA |
| Father | 53 | M | DOE | Sigmoid septum, no obstruction | NSR, RVH | 19 | 82 | None | BBs |
DOE: Dyspnea on Exertion; RV: Right Ventricular; AF: Atrial Fibrillation; PAF: Paroxysmal Atrial Fibrillation; LBBB: Left Bundle Branch Block; RBBB: Right bundle Branch Block; NSR: Normal Sinus Rhythm; NSST-T: Non-Specific ST-T; ASA: Alcohol Septal Ablation; LVOT: Left Ventricular Outflow Tract; RVH: Right Ventricular Hypertrophy; BAV: Atrioventricular Block; BBs: Beta-Blocker agents; ICD: Implantable Cardioverter-Defibrillation; M: Male; F: Female. *Novel mutation in TNNT2 gene.
Table 2: Demographic, clinical, ECG and echocardiography characteristics of patients having genotype positive and their relatives.
Here we identified a novel variants not previously reported in patients with Hypertrophic Cardiomyopathy, c.822-2A>G on TNNT2. The variants happen at a splice-site; thus, the end result would create an aberrant splicing of TNNT2, leading to nonsense mediated decay of the RNA product, reducing the total Cardiac muscle troponin T end-product, thus leading to the phenotypes of these patients.
Synopsis of the novel TNNT2 pathogenic variant
Figure 1 shows the pedigree of the family with the novel c.822- 2A>G variant in TNNT2 gene. The proband (subject IV.4) was a 25-year-old woman who had dyspnea on exertion and chest pain. Echocardiography showed the whole left ventricular hypertrophy involving septum, anterior, anterolateral walls, and also apex with the severe LV outflow tract obstruction (pressure gradient, 110 mmHg). She was treated with Alcohol Septal Ablation (ASA) and Beta-Blocker Agents (BBs), the symptoms were alleviated.
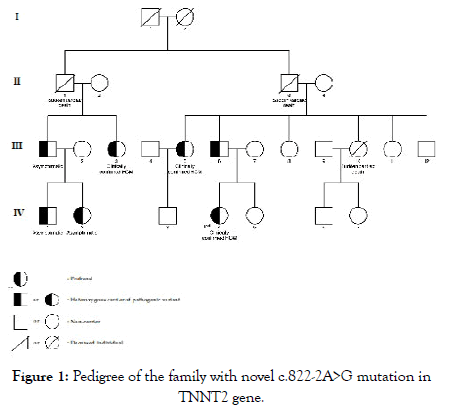
Figure 1: Pedigree of the family with novel c.822-2A>G mutation in TNNT2 gene.
At the time of research, 11 of her relatives were screened for HCM. Three of them including the proband’s father (subject III.6) and 2 of her aunts (subject III.3 and subject III.5) were clinically confirmed to have HCM and also carrying the same pathogenic variant as the proband. Meanwhile, her uncle (subject III.1) and his children (subject IV.1, IV.2) who were also the carrier of this variant had no clear indication of HCM. The patient’s mother (subject III.7), her sister (subject IV.5), two of her aunts (subject III.8 and III.11), and her uncle (subject III.12) did not have any manifestations of HCM and were genetic negative confirming by using Sanger sequencing method.
Three other relatives (subject II.1, II.3, and III.10) were reported to suffer Sudden Cardiac Death (SCD) at the age of 40, 63 and 30, respectively. All of them were asymptomatic and did not have a history of any diseases prior to their death. Clinical characteristics of subjects carrying the c.822-2A>G variant in TNNT2 gene were shown in Table 2.
In this study, HCM-causative variants of four genes including MYH7, MYBPC3, TNNT2 and TNNI3 which encoded for betamyosin heavy chain, myosin binding protein C, cardiac troponin T and cardiac troponin I, respectively found to be associated with HCM in our cohort. Among 41 individuals from 10 patient families, there were 5 out of 10 patients (50%) carried a single variant related to MYH7, MYBPC3, and TNNT2 genes and 11 (35.5%) of their relatives bringing the same variants. A novel c.822- 2A>G variant of the VUS splicing receptor zone of TNNT2 was found in a family with 7 individuals positive with HCM genetic testing but only 4 out of these 7 individuals had HCM phenotype.
The prevalence of attributable pathogenic variants in our cohort having familial HCM was lower than the reported prevalence in the US and French cohort, which were 54.2% and 60.6% [4,12] but higher than the other cohorts in Portugal, Taiwan, Finland and Japan, which accounted for 28%, 34.2%, 38.2% and 43.8% [13- 16], respectively. Recently, a study in Vietnamese HCM patients in the South of Vietnam, in which a panel of 23 HCM-related genes pointed out that 43.4% of the patients diagnosed HCM carried variants mostly focused on MYH7, TPM1 and TNNT2 genes which were linked with more severe clinical manifestations [8].
Many previous studies in patients with familial HCM in the Caucasian population and Asian population found mostly variants of MYBPC3 and MYH7. Our study noted that in two family’s which carried pathogenic variants of MYH7 gene (p.T1377M and p.R782H), sudden cardiac deaths of family member occur more frequently. Fujita E et al. reported Japanese patients with HCM carrying variants in TNNT2 gene that associated with the risk of sudden cardiac death in youth. In our cohort, family number 3 was found to carry the novel variant c.822-2A>G in TNNT2 gene, which might cause SCD for 3 different family members. In this family, there are 6 family members carrying the same c.822-2A>G variant with the patient, but only 3 of them expressed the HCM phenotype, including the patient’s father and her 2 aunts. Three other individuals were genotype positive-phenotype negative, but 2 of them (subject IV.1, IV.2 in figure 1) were under the age of 5.
The gene we chose to investigate in our panel was the more frequently reported to be associated with HCM, and each has a direct role in the heart muscle constitution and function. However, we suspect that there would be other potentially novel pathogenic gene or pathogenic variants in the non-coding region which could be identified with an expanded panel of the Whole Exome or Whole Genome sequencing. In this study due to the number of cases and short observation time, we were unable to gauge the difference in disease severity of HCM in relation to each diseasecausing gene. Future studies would also focus on prospective followup of the patient to assess the natural history of disease in relation to their pathogenic variants.
This study was conducted on a group of 10 probands with HCM and their family members. A targeted Next-Generation sequencing panel was utilized and in-depth clinical profiling was made to study the prevalence of genetic aberration and to study the genotypephenotype correlation in this group of patients. This case series highlights the importance of genetic testing for clinically confirmed HCM patients and family members. The genetic information can be used as a molecular marker to complement the clinical presentation in the diagnosis of HCM, as well as a prognosis tool for the patients and their family members.
Ethic approval and consent to participate
The study design was reviewed and approved by the Ethical board of Hanoi Medical University. The study complies with the Declaration of Helsinki regarding the use of human samples and identifiable information. Informed consent was obtained from the patients regarding the use of the samples and information for research purposes.
Consent to publish
The patients gave consent to publish the patients’ information, including clinical and genetics information. No identifiable information was disclosed in any form.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. We however cannot provide personal information or data contain identification of the patients in any form.
Competing interests
The authors declare no conflict of interest.
Author Contributions
Author Hung Manh Pham, Van Khanh Tran, Trung Anh Mai, and Long Hoang Luong conceived and design the study and analysis. Author Thanh Van Ta and Huy Thinh Tran contributed to the development of the study’s idea and design. Author Hoai Thu Nguyen Thi, May Le Pham, and Chi Khanh Nguyen contributed to data collection and carry out the experiments. Author Minh Nhat Pham, Can Thuy Do and Thanh Tuan Le performed analysis and finalizing the results. Author Hung Manh Pham, Van Khanh Tran, Long Hoang Luong, and Trung Anh Mai contributed to the drafting of the manuscript. All authors have read and approved of the final version for publication.
We sincerely thank to the patients and their parents for giving us consents and allowing us to publish the data. We thank our colleagues at who help us in patient recruitment and clinical data collection.
Citation: Pham HM, Tran VK, Mai TA, Luong LH, Pham ML, Nguyen CK, et al. (2021) A Case Series of Hypertrophic Cardiomyopathy in Vietnam Reveal Novel Pathogenic Variants in Tnnt2 Gene. J Med Diagn Meth. 10:321.
Received: 16-Mar-2021 Accepted: 30-Mar-2021 Published: 06-Apr-2021 , DOI: 10.35248/2168-9784.21.10.321
Copyright: © 2021 Pham HM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.