Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2024)Volume 17, Issue 2
The FBN gene family is essential to many biological processes, including plant development and response to a variety of biotic and abiotic stresses. However, there is currently no information about this gene family in maize. In this study, we performed a genome-wide study of the FBN gene family in maize, identified 6 ZmFBN genes by a different bioinformatics method, and further characterized them to understand their structure and function. For this purpose, genome-wide analysis is performed to identify the non-redundant FBN genes in the genome of maize. This research aims to explore the function of FBN genes in maize. Genomic and CDS sequences were used to reveal structural features. The evolutionary analysis was performed by constructing a phylogenetic tree. Synonymous and non-synonymous (Ka/Ks) ratios were also calculated for ZmFBN. Gene expression analysis was performed by using the NCBI GEO dataset and a heat map generated to identify the abiotic stress genes of FBN in maize. According to our results one out of six members ZmFBN3 (Zm00001d002384) of FBN gene family was regulated under stress condition, and supported the maize plant under abiotic condition.
Maize; FBN gene; Grain yield; Abiotic stress; Gene expression
The most frequently cultivated crop in the world is maize (Z. mays). Its reaction to numerous environmental stress factors is highly dynamic and complex in nature. On the other hand, climate change is predicted to intensify and spread biotic and abiotic stress factors more frequently. 80% of human food is produced by crops, with grains contributing to 50% of the world's food supply [1]. It provides the continually increasing human population by fulfilling their nutritional requirements either directly as a food for human consumption or indirectly as feed for livestock. Furthermore, due to its ability to produce bio-ethanol, maize, referred to as corn in the United States, serves as an environmentally friendly substitute to fossil fuels [2]. Because changing climatic conditions increase variability in maize production, it is essential to evaluate the overall impact of climate change on the growth and development of this staple crop in order to estimate its consumption. Temperature extremes, such as drought, nutrient shortage and minerals are among the various abiotic stresses that are predicted to have the biggest effect on the yield of maize overall. The two most significant climate variables, based on recent research, temperature and precipitation, with radiation additionally performing a significant role in yield. However, the current climate of droughts, temperature extremes and waterlogging has significantly lowered maize development thus decreasing its yield [3]. Due to drought or waterlogging, every year loses in maize yield [4].
Members of the fibrillin family play a role in a variety of functions including growth, development and resistance against abiotic stress. The most common type of protein found in chloroplast Plastoglobules (PGs) is a fibrillin (FBN) protein, which is plastid lipid-associated and highly conserved [5]. In plastid stability, plant growth and development, and stress response, FBNs have essential regulatory functions [6]. Widely present in photosynthetic species, including cyanobacteria and plants, FBN proteins have a conserved Plastid-lipids Associated Protein (PAP) domain [7,8].
In many processes including plastid structure stabilization, organ development, stress response and hormonal signal transmission, the FBN gene family has demonstrated important functions [9].
Many biological functions, including stress response, are significantly regulated by genes of the FBN family. FBN family genes in maize, yet, remain unknown and no comprehensive study of this family in maize has been published. In this study, bioinformatics tools were used to analyze the phylogenetic relationship, conserved domain, collinearity and cis-acting element in the ZmFBN gene family promoter regions. The results of this study, helps in future research into the biological function of the FBN gene during various phases of maize development.
ZmFBN gene family identification in maize and other crop
To identify all sequences related to FBN family, from TAIR database reference gene AtFBN was downloaded. Identified domain was BLAST in maize Genomic Database (GDB) to get putative genes of FBN gene family in maize. The domain was also blasted to find the putative FBN genes in A. thaliana, T. aestivum, G. hirsutum, A. comosus, B. rapa, G. max and P. trichocarpa. All the sequences were downloaded from Phytozome v13.
Protein motif analysis of ZmFBN
The motif analysis will be performed by using ZmFBN protein sequences on MEME. It will generate an XML file as result. The graphical presentation and sequence logos of each motif will appear on the result page. The graphic representation of motifs will also be downloaded TBtools software by using a MEME XML file.
Conserved domain analysis of ZmFBN
In each sequence, conserved domains were identified by submitting ZmFBN peptide sequences on the Conserved Domains Database (CDD) tool of National Center for Biotechnology Information (NCBI) and the generated CDD file used as input in TBtool software to generate its graphical structure. The CDD tool help in determining the molecular function of the protein [10,11].
Gene structure analysis of ZmFBN
Genomic and Complementary DNA Sequences (CDS) will be retrieved from https://gamma.maizegdb.org and the obtained sequences will be subjected to Gene Structure Display (GSD) server 2.0 for the development of the gene structure of ZmFBN.
Sequence logos and phylogenetic analysis
The peptide sequences of Arabidopsis thaliana and Z. mays were aligned by MEGA11. By using CLustal W all sequences were aligned and the structure was constructed using the TBtool software. For comparative phylogenetic and evolutionary analyses of FBN genes, downloaded and aligned by CLustal W, the FBN peptide sequences of maize and other crops were used to create a phylogenetic tree that used the neighbor-joining method to estimate the evolutionary history [12].
Chromosomal distribution, prediction of Ka and Ks ration and synteny analysis
To create the chromosomal length file, the maize genome assembly file will be used. Physicochemical properties data were used to perfumed chromosomal distribution analysis. The online tool PhenoGram will be used for chromosomal analysis. It will produce plots that depict where genes are located on chromosomes. Website selected is http://visualization.ritchielab.org/phenograms/plot as it is a versatile and user-friendly tool. The created pairings were processed through the TBtool program to calculate the rates of synonymous and non-synonymous substitution (Ka). The type of codon selection that resulted during evolution was further determined using the Ks/Ka ratio [13]. Further, using the formula T=Ks/2 and assuming a clock rate of (2*6.1*10^-9) substitutions/synonymous site/year for maize, the approximate period of duplication event was calculated. For collinearity analysis, the genome and General Feature Format (GFF) 3 files of maize and rice were used. Required files generated using one step MCscan. The results were visualized using TBtool software [14].
Sub-cellular localization, physicochemical properties and cis acting elements analysis of ZmFBN genes
By CELLO, Subcellular Localization Predictive System (SLPS) and WoLF PSORT subcellular localization was predicted. The physicochemical properties of ZmFBN genes were also identified by using Phytozome v13 and the Expasy Protparam tool. It describes the chromosome number, base pairs of peptides, CDS, transcript and genomic sequence, the molecular weight of protein and the number of introns and exons. For cis-element analysis of FBN genes, promoter regions ~ 2 kb upstream sequence region of the start codon were retrieved from Phytozome v13 and subjected to the PlantCare database for the identification of binding sites for the known transcription factor [15].
Protein-protein interaction network of ZmFBN
The structure prediction analysis has performed by using ZmFBNs protein sequences. Structures of all the FBNs were predicted through the online website https://yanglab.nankai.edu.cn/trRosetta/. To predict the Protein-Protein Interaction (PPI) of ZmFBN genes, protein sequences were subjected to the STRING search portal for the determination of PPI. A biological database and online resource for protein-protein interactions, known as STRING.
Gene expression profile of ZmFBN genes
For in-silico expression analysis of ZmFBN genes at different stages, transcriptional data of the genes were obtained from NCBI GEO DataSet. A heat map was produced by using TBtool software to identify the yield and quality controlling genes. All the analysis helped in the identification and characterization of the FBN gene family in maize.
Identification and classification of ZmFBN genes
Through the use of the BLAST method, 6 ZmFBN genes were found in maize in the present research. The results provided basic information on the gene familial, CDS lengths, protein lengths, amino acid counts and molecular weights as well as their equipotential points, aliphatic indexes and GRAVY values as shown in Table 1. The protein length varied from 528 bp to 5857 bp in ZmFBN gene family, and number of exons in genes ZmFBN1 to ZmFBN6 ranges from 1 to 10. ZmFBN6 are hydrophobic proteins, according to the expected average hydrophilic coefficient (GRAVY), while others are hydrophilic proteins. Proteins found in the ZmFBN gees ranged from 176 bp to 361 bp amino acids in length from ZmFBN1 to ZmFBN6. Gene characteristics of ZmFBN genes, including Coding Sequence (CDS) length varied from 528 bp to 1084 bp, the molecular weight predicted for these proteins ranged from 18828.75 to 39237.95 kDa in ZmFBN genes respectively (Table 1).
| Gene ID | Gene name | Genomic sequence | Transcription sequence | CDS bp | Peptide bp | PI | Gravity | Molecular weight | Amino acid |
|---|---|---|---|---|---|---|---|---|---|
| Zm00001d021351 | ZmFBN1 | 5857 | 1458 | 1084 | 361 | 8.7 | -0.062 | 39237.95 | 360 |
| Zm00001d025574 | ZmFBN2 | 3591 | 1475 | 945 | 315 | 5.9 | -0.141 | 33610.25 | 314 |
| Zm00001d002384 | ZmFBN3 | 1994 | 1495 | 750 | 250 | 9.2 | -0.081 | 27194.41 | 249 |
| Zm00001d003470 | ZmFBN4 | 5260 | 1498 | 957 | 319 | 5.4 | -0.142 | 34280.94 | 318 |
| Zm00001d033666 | ZmFBN5 | 3767 | 1278 | 864 | 288 | 9.9 | -0.277 | 31115.86 | 287 |
| Zm00001d018348 | ZmFBN6 | 528 | 528 | 528 | 176 | 10 | 0.185 | 18828.75 | 175 |
Note: CDS: Coding Sequences; bp: Base pairs; PI: Isoelectric Point
Table 1: Basic information of FBNs gene family in Z. mays.
Conserved domain, protein motif, gene structure and cis acting elements of ZmFBN genes
Exon/intron distribution patterns of genes have a relationship with their biological functions and the evolutionary relations between different species of plants. The conserved gene structure and domain analysis was analyzed by GSDS server 2.0 and CDD tool of NCBI. The results showed that ZmFBN family is divided into two subfamilies with typical subfamily domain. The PAP fibrillin (PF04755) domain was present in all motifs contained the fibrillin which is plastid-lipids associated protein that plant growth and development, as well as plant response to both abiotic and biotic stress. The results indicated that the PAP fibrillin domain was present in two genes and PAP fibrillin super family was present in all genes as shown in Figure 1 and number of exons in genes ZmFBN1 to ZmFBN6 ranges from 1 to 10. One gene has no introns (ZmFBN6). The five of the six genes analyzed in this study contain introns. The two genes (ZmFBN4 and ZmFBN2) contained the two intron and three exons, one gene (ZmFBN2) have three introns and four exons and one gene (ZmFBN1) have nine introns and ten exons as shown in Figure 2. MEME analysis was used to determine the putative motifs of the FBN family in maize and all members contained one different motif for each subfamily, verifying that they belonged to the same subfamily as shown in Figure 3. The results showed that most of the members that are closely linked share common patterns. Plant growth, development and acceptability to biotic and abiotic stress were predicted functions of the cis acting essential components in the maize region. such as ABRE (involved in abscisic acid) and GARE (involved in gibberellins) and AUXR (involved in auxin responsiveness) to environmental challenges would increase in part because such stresses would trigger the activation of expansion genes with their responsive cis-acting elements. The gibberellin responsive elements were most abundant in the promoters of the ZmFBN2 gene. The Light Response (LR) elements are highest in ZmFBN1 and AUX and MYB elements are lowest in this gene. The MYB element was present only in one gene (ZmFBN1). The AUXNR was lowest in ZmFBN1 and ZmFBN3 genes and MeJAR elements were not present in ZmFBN3 gene (Figures 1-4a).
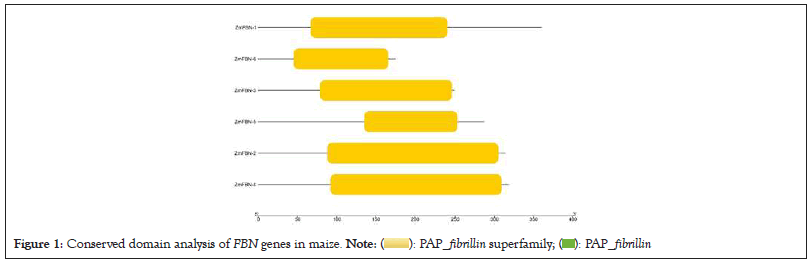
Figure 1: Conserved domain analysis of FBN genes in maize. 
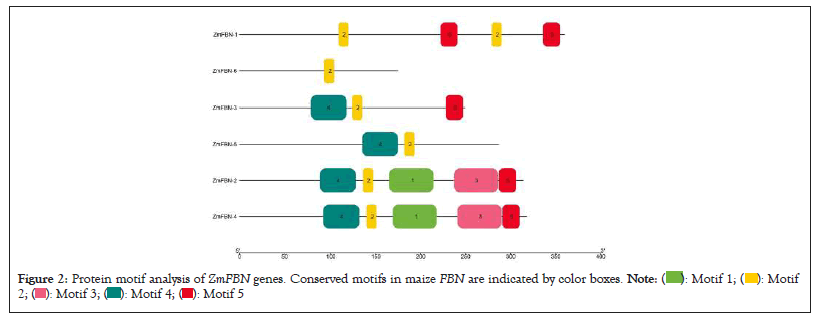
Figure 2: Protein motif analysis of ZmFBN genes. Conserved motifs in maize FBN are indicated by color boxes. 
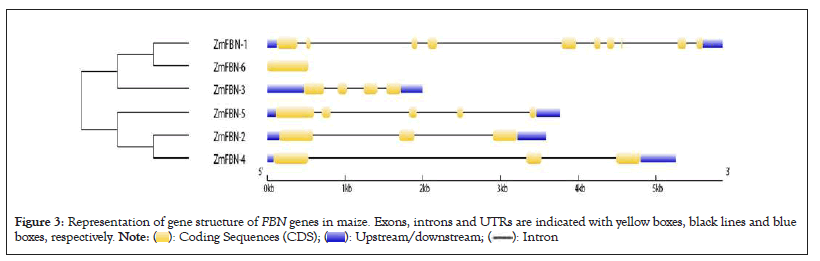
Figure 3: Representation of gene structure of FBN genes in maize. Exons, introns and UTRs are indicated with yellow boxes, black lines and blue boxes, respectively. 
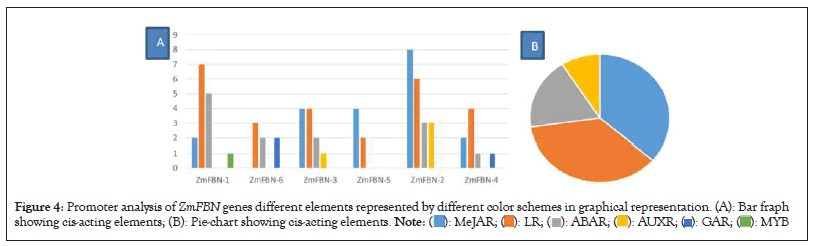
Figure 4: Promoter analysis of ZmFBN genes different elements represented by different color schemes in graphical representation. (A): Bar fraph showing cis-acting elements; (B): Pie-chart showing cis-acting elements. 
Phylogenetic analysis, sequence logo, sub-cellular localization and protein interaction analysis
The 6 member of FBN genes in maize and divided into two groups. The red color indicates group1 and blue color group2 as shown in Figure 5a. To better understand the evolutionary relationships of the FBN family, a phylogenetic tree was constructed for 90 members of this family in seven different species, including OsFBN, AtFBN, GhFBN, GmFBN, AcFBN, PtFBN and BrFBN proteins as shown in Figure 5b. According to the phylogenetic tree, FBN family members can be classified into five groups, including group 1, group 2, group 3, group 4 and group 5. The group 3 contained 3 members, group 4 with 1 and group 5 contained 2 members of ZmFBN gene family. The phylostarum analysis of FBN gene family identified the earliest plant lineage. Further, the FBN genes were present in G. hirsutum, G. max and P. trichocarpa, dicotyledons (B. rapa), A. comosus (angiosperm), monocotyledons (B. distachyon) dicots (A. thaliana and monocots (Z. mays). The results of this study showed that the FBN gene family originated in phylostratum, an early land plant and that FBN orthologous genes may exist through the plant kingdom. These findings showed that the genes for FBN came from early land plants called phylostratum and that the plant kingdom contains probable orthologous genes for FBN (Figures 5a and 5b and 6).
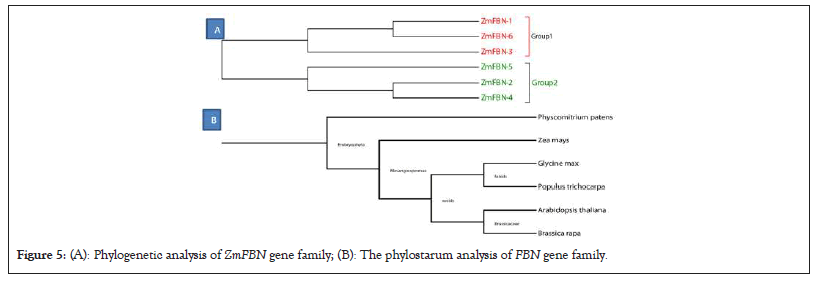
Figure 5: (A): Phylogenetic analysis of ZmFBN gene family; (B): The phylostarum analysis of FBN gene family.
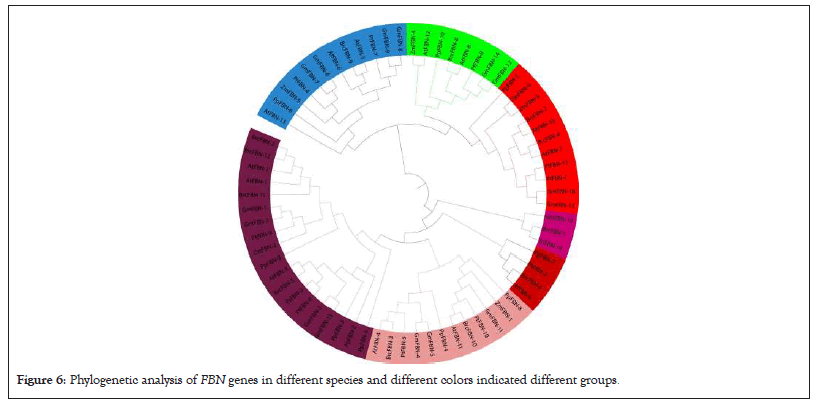
Figure 6: Phylogenetic analysis of FBN genes in different species and different colors indicated different groups.
The subcellular localization of proteins using CELLO (Subcellular Localization Predictive System) results predicted that ZmFBN1 is located in outer membrane with variability value (2.500), ZmFBN2 is located in outer membrane with variability value (2.625), ZmFBN3 is located in periplasmic with variability value (1.996), ZmFBN4 is located in outer membrane with variability value (2.462), ZmFBN5 is located in periplasmic with variability value (3.121) and ZmFBN6 is located in periplasmic with variability value (1.805) as shown in Figures 6 and 7. Using WoLF PSORT, proteins' subcellular localization (Protein Subcellular Localization Prediction) predicted that ZmFBN located in outer membrane and periplasmic membrane as shown in Table 2. Sequence logos provide more specific information regarding sequence similarities, significant alignment characteristics and patterns of sequence conservation. The sequence logos of Arabidopsis and maize are showing divergence. Our results showed that the FBN gene family among all species is not conserved (Figures 7 and 8 and Table 2).
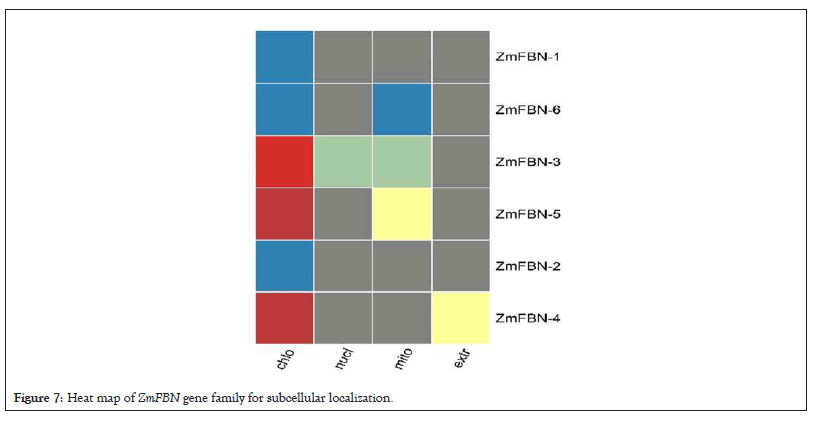
Figure 7: Heat map of ZmFBN gene family for subcellular localization.
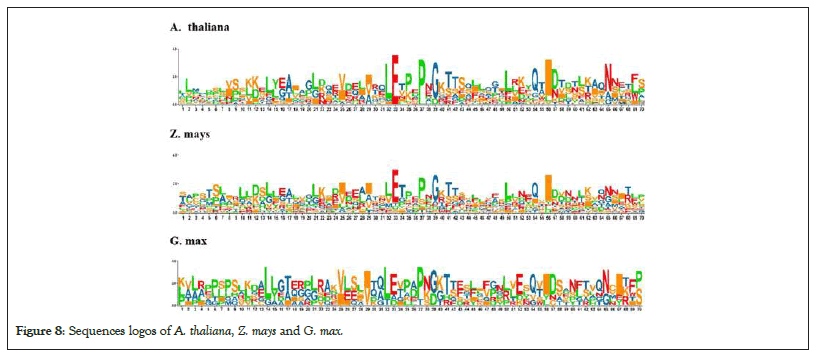
Figure 8: Sequences logos of A. thaliana, Z. mays and G. max.
| Gene Ids | Chloroplast | Nucleus | Mitochondria | Extracellular matrix | Sub-cellular localization (significant) |
|---|---|---|---|---|---|
| ZmFBN1 | 14 | - | - | - | Outer membrane |
| ZmFBN2 | 14 | - | - | - | Outer membrane |
| ZmFBN3 | 12 | 1 | 1 | - | Periplasmic |
| ZmFBN4 | 13 | - | - | 1 | Outer membrane |
| ZmFBN5 | 10 | - | 4 | - | Periplasmic |
| ZmFBN6 | 7 | - | 7 | - | Periplasmic |
Table 2: Prediction of subcellular localization of FBN protein through Wolf PSORT.
To understand the functions and metabolic pathways of the ZmFBN members, the protein-protein interaction network was predicted with the online String database as shown in Figure 9. Analysis predicted that protein ZmFBN has interaction with other proteins. Protein structure of FBN gens was restrained Rosetta (trRosetta) online tool as shown in Figure 10. The protein ZmFBN4 showed interaction with APE1, ZmFBN1, and GRMZM5G359786. The ZmFBN2 showed highest interaction with ZmFBN4, GRMZM5G359786, GRMZM5G124473 and ZmFBN3. The protein structure was determined for six ZmFBNs representative proteins, which shared the 90% similarities with each other. This level of structural similarity was adequate for analysis protein structures (Figures 9 and 10).
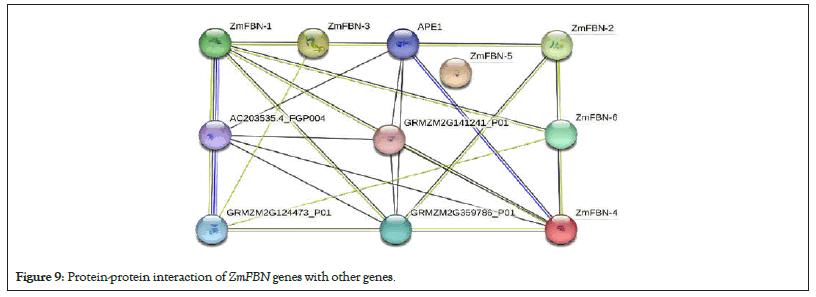
Figure 9: Protein-protein interaction of ZmFBN genes with other genes.
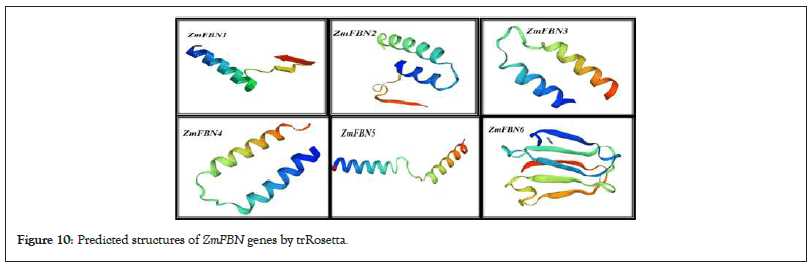
Figure 10: Predicted structures of ZmFBN genes by trRosetta.
Chromosomal distribution, prediction of Ka and Ks ration and synteny analysis
The 6 FBN genes were distributed on five maize chromosomes on based of genomic data. ZmFBN5 gene were located on the chromosome 1, ZmFBN3 and ZmFBN4 genes were located on the chromosome 2, ZmFBN6 on chromosome 5, ZmFBN1 was located on chromosome 7 and ZmFBN2 gene was located on chromosome 10 as shown in Figure 11. The ZmFBN phylogenetic tree revealed only 2 gene pair (ZmFBN1, ZmFBN6 and ZmFBN2, ZmFBN4). The synonymous and non-synonymous (Ka/Ks) value of this gene pair was calculated to determine the extent and nature of selection pressure on the duplicated gene pair. The calculated Ka/Ks value=0.24608463. In this study, we found that ZmFBN genes showed Ka/Ks values greater than 1. The Ka and Ks value revealed that for 1 pair of segmental gene was present in maize as shown in Table 3. The synteny between species is useful for understanding how and why specific gene families and functions have evolved over time. The FBN genes in maize share homologous pairs with FBN genes in other plants, suggesting that FBN may have been crucial to the evolution of Z. mays. There were collinear gene pairs in Z. mays and O. sativa. 1.0 represented collinearity between chromosome 1 in Z. mays (ZmFBN5) and chromosome 3 in O.sativa (OsFBN1). The 2.0 represented collinearity between chromosome 2 in Z. mays (ZmFBN3) and chromosome 4 in O. sativa (OsFBN2). The 7.0 represented collinearity between chromosome 7 in Z. mays (ZmFBN1) and chromosome 7 in O. sativa (OsFBN3) (Figures 11 and 12 and Table 3).
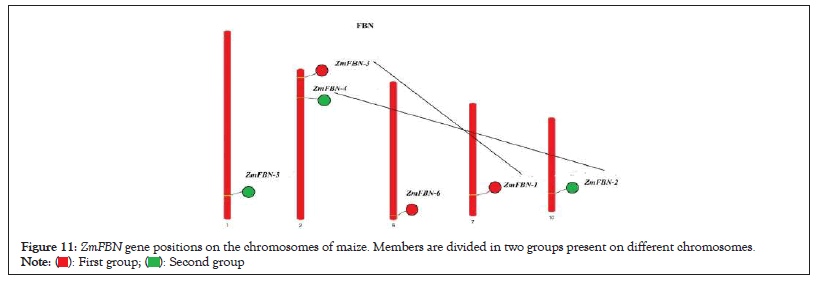
Figure 11: ZmFBN gene positions on the chromosomes of maize. Members are divided in two groups present on different chromosomes. 
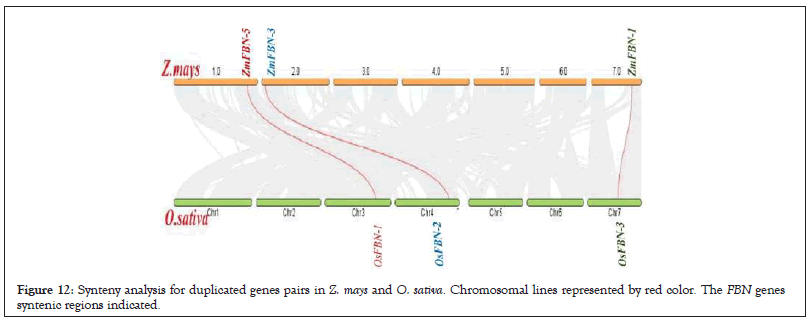
Figure 12: Synteny analysis for duplicated genes pairs in Z. mays and O. sativa. Chromosomal lines represented by red color. The FBN genes syntenic regions indicated.
| Gene-1 | Gene-2 | Ka | Ks | Ka/ks | Time (MYA) | Duplication | Selection pressure |
|---|---|---|---|---|---|---|---|
| ZmFBN2 | ZmFBN4 | 0.05120049 | 0.2080604 | 0.24608463 | 4 | Segmental | Positive |
Note: Ka: Non-synonymous; ks: Synonymous
Table 3: Synonymous (Ks) and non-synonymous (Ka) calculation.
Gene expression analysis of ZmFBN genes
For gene expression analysis Gene Expression Omnibus (GEO) dataset was used. The high throughput sequencing for expression profiling. A group of hormones known as Brassinosteroids (BRs) are unique to plants and serve important functions in their physiology as well as the control regarding the way they react to stress. The effects of 24-epibrassinolide (EBL), a synthetic BR, on hydroponically grown maize seedlings were studied in this research using a combined physiological and molecular approach. To further examine the effects of this chemical on shoot growth, a 48-hour exposure period and a treatment concentration of 1 nM EBL were used. RNA sequencing was used to study the root gravitropic response. Exogenous management of EBL during the seedling stage resulted in a considerable decrease in root and shoot length, according to our results. The highest concentrations (10 and 100 nM) severely inhibited primary root growth (10% and 25%, respectively) after 24 hours. After 24 hours, the lowest concentration (1 nM EBL) showed no effect on PR formation. However, after 48 hours of treatment, the impacts on root growth were visible even at the lowest dose (1 nM) [16]. The expression of all FBNs in both leaf control, leaf EBL, shoot control and shoot EBL samples on vegetative and reproductive stages. Gene expression analysis revealed that one out of six members ZmFBN3 gene was highly regulated under both leaf control, leaf EBL, shoot control and shoot EBL conditions and supported the maize plant under stress condition (Figure 13).
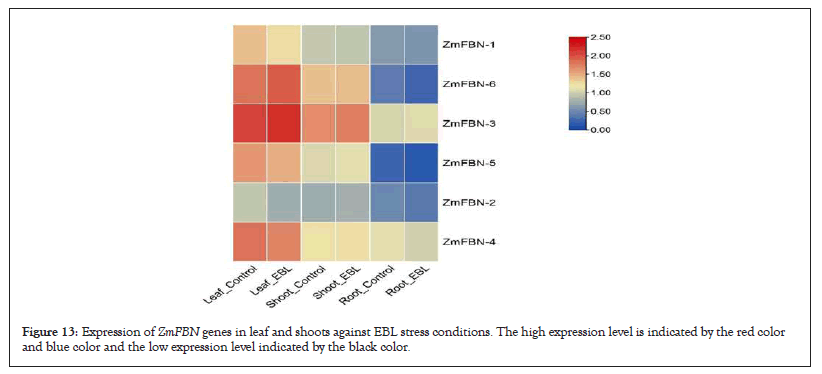
Figure 13: Expression of ZmFBN genes in leaf and shoots against EBL stress conditions. The high expression level is indicated by the red color and blue color and the low expression level indicated by the black color.
As a unique transcription factor family in plants, the FBN gene family is essential for controlling plant growth, physiological functions, and stress response. Fibrillins (FBNs) are named after fibrils since these proteins were discovered in fibrils in dog rose chromoplasts (Rosa rugosa) and bell pepper (Capsicum annuum) fruit [17]. The creation of lipoprotein structures, photosynthesis and reactions to both abiotic and biotic stress are only a few of the crucial biological roles that FBN proteins play.
Conserved domains, motif patterns, gene chromosomal distributions, phylogenetic relationships, gene structures, synteny analyses and protein-protein structure analysis have been studied with bioinformatics in the present study. ZmFBN proteins were found in the periplasmic and outer membranes of cells based on predicted subcellular locations, indicating that these genes function as transcription factors.
ZmFBN genes in the same subgroup typically have intron and exon positions that are comparable, indicating that their functions are similar. One gene has no introns (ZmFBN6). The two genes (ZmFBN4 and ZmFBN2) contained the two intron and three exons, one gene (ZmFBN2) have three introns and four exons and one gene (ZmFBN1) have nine introns and ten exons.
The majority of ZmFBN proteins belonging to the same subfamily typically include similar motifs, according to the conserved motif analysis. The domains that we found can be divided into two groups. For ZmFBN members, the PAP family and PAP superfamilies are two of the most important domain groups. These ZmFBN genes are important in the response to environmental stress, according to the promoter analysis. The FBN family has been analyzed for synteny between maize and rice, and it was shown that the collinearity blocks between FBN members of the PAP subfamily were the highest. These findings point to segmental duplication events in the evolution of the FBN family, which allowed for its further expansion. Furthermore, fibril-like structures can be produced in vitro using a mixture of bicyclic carotenoids, lipids and FBN protein [18]. The most common proteins in chloroplast PGs are fibrillin (FBN) proteins, which are plastid lipid-associated and highly conserved proteins that are encoded by nucleus genes. Plant growth and development, plastid stability, and stress response all showed significant regulatory roles for FBNs.
The results may provide some helpful guidelines for developing new maize varieties with high susceptibility to develop in stress condition and also provided the theoretical basis of FBN-domain for agricultural applications. Although, this study might make easier to understand how FBN genes work in maize and other crops.
In this study, 6 FBN gene family members were investigated. We analyzed ZmFBNs using bioinformatics techniques such as gene identification, sequence logo, gene expression analysis phylogenetic analysis, chromosomal placement, interaction protein network analysis, cis-regulatory elements and gene structure. Determining the mechanism of stress management is also challenging. In-silico analysis provides useful information for future functional investigations in stress biology considering the fact that stress control is a complex system. The expression profile data also indicate that ZmFBN were responsive to numerous biotic and abiotic stressors. As a result, this study provides functional information on FBN genes to help improve plant growth, stress tolerance, and to help us comprehend the many developmental processes in maize.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Bashir S, Farid B, Zafar MZ, Rehman SU, Shah MR, Atta Z (2024) A Comprehensive Study of ZmFBN Gene Family and their Biotic and Abiotic Stress Response in Maize. J Proteomics Bioinform.17:668
Received: 14-Aug-2023, Manuscript No. JPB-23-26079; Editor assigned: 16-Aug-2023, Pre QC No. JPB-23-26079 (PQ); Reviewed: 30-Aug-2023, QC No. JPB-23-26079; Revised: 06-Sep-2023, Manuscript No. JPB-23-26079 (R); Published: 24-Jun-2024 , DOI: 10.35248/0974-276X.24.17.668
Copyright: © 2024 Bashir S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.