
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research - (2022)Volume 12, Issue 5
The accurate detection of leukocytes is the basis for the diagnosis of blood system diseases. However, current methods and instruments either fail to fully automate the identification process or have low performance. To improve the current status, we do need to develop more intelligent methods. In this paper, we investigate fulfilling high-performance automatic detection for leukocytes using a deep learning-based method. A complete working pipeline for building a leukocyte detector is presented, which includes data collection, model training, inference, and evaluation. We established a new leukocyte dataset that contains 6273 images (8595 leukocytes), considering nine common clinical interference factors. Based on the dataset, the performance evaluation of six mainstream detection models is carried out, and a more robust ensemble scheme is proposed. The mAP@ IoU=0.50:0.95 and mAR@IoU=0.50:0.95 of the ensemble scheme on the test set are 0.853 and 0.922, respectively. The detection performance of poor-quality images is robust. For the first time, it is found that the ensemble scheme yields an accuracy of 98.84% for detecting incomplete leukocytes. In addition, we also compared the test results of different models and found multiple identical false detections of the models, then provided correct suggestions for the clinic.
Detection algorithm; Deep learning; Ensemble; Leukocyte
It is of great significance for clinicians to recognize peripheral blood leukocytes through blood smears for the diagnosis of blood cancer, and the automation of this process can be a great help in the clinic. The blood smear suggests a possible diagnosis in diagnosis of leukemia, especially important for clinical detection of Burkitt lymphoma and acute promyelocytic leukemia because it facilitates rapid diagnosis and timely treatment [1]. However, this is a complicated, time-consuming, laborious, and subjectively influenced work by the doctor. At the same time, the observer is required to have sufficient experience [2]. Therefore, it is very necessary to study a computer-aided system for automatic detection of peripheral blood leukocytes with high accuracy.
In the past, the research community and medical industry have attempted to automate the detection of leukocytes, and this automation has become a developmental trend in medical examination for blood cells [3]. In the medical industry, there are several Automated Cell Morphology (ACM) systems. For example, Cella-Vision [4] fulfills some automation with digital imaging technologies, and MED-ICA EasyCell® Assistant [5] uses image processing and pattern recognition technologies. However, these instruments are based on traditional machine learning methods [6]. Compared with clinical experts, although these methods provide useful assistance and can accelerate the process of recognizing blood cells, their performance is still far behind the human experts’ level, and cannot reliably work independently [6,7].
Since AlexNet excelled in the 2012 ImageNet competition, deep learning technology showed a promising solution in medical image application [8]. Since then, a large number of publications [9-13] have reported that the Convolutional Neural Networks (CNN) model, i.e., deep learning, is competent for image recognition tasks in different areas. Thanks to the unified homogenous model of CNN, making use of it avoids the disadvantages of multi-step traditional machine learning methods. Recently, different studies have improved algorithms from different aspects, which have indeed improved the accuracy of identifying leukocytes, but these deep learning methods [2,14,15] focus on leukocyte classification rather than leukocyte detection. Some restrictive assumptions are imposed on the classification task and the dataset. Leukocyte classification requires that the segmentation must have been done, the existence of target cells in the image must be guaranteed, and the images mostly contain only one leukocyte. This segmentation breaks the automatic process and causes inconvenience in practical clinical application. Furthermore, to our best knowledge, the 13 public datasets retrieved are all for leukocytes classification research [16-25], which all contain many images with only one leukocyte.
Some recent research efforts have focused on leukocyte recognition as multi-object detection. The multi-object detection method can automatically locate the objects and determine their types as well, dropping the restrictive assumptions of the classification task. Compared with leukocyte classification, the degree of automation is higher in terms of process: the quantity, locations, and types of leukocytes can be obtained simultaneously, and the high accuracy rate of deep learning for identifying leukocytes is maintained. However, the current research on the detection of leukocytes uses a relatively single type of leukocyte in the dataset, and it is difficult to evaluate the level of recognition of the five types of leukocytes [26]. Moreover, the multi-center problem of the dataset is not considered [27,28], and the generalization of the solution is not adequately discussed. In other words, the detection performance on new data collected from other hospitals needs further study. Furthermore, the public leukocyte dataset found so far not enough to support the detection of leukocytes [16-25].
Therefore, we collected data from multiple hospitals and established a dataset suitable for the detection of leukocytes for the first time, which considered the nine interference factors that are likely to affect the performance of the detectors in detecting leukocytes [29] in an attempt to fundamentally solve the multicenter heterogeneity problem. Based on the dataset, we tested the performance of six mainstream detection models and then tried to propose a new and more robust model using an ensemble scheme.
Working pipeline of building up an AI-based detector
To apply AI-based detection method onto automatic identification of leukocytes, the working pipeline of constructing a deep learningbased detector consists of 4 stages: 1) data preprocessing; 2) model training; 3) inference; and 4) evaluation.
Data preprocessing: The proposed leukocyte dataset is preprocessed in two different data formats using dataset conversion toolbox developed by the authors. The processed the dataset includes ground truth labels and split subsets in both VOC style format and COCO style format. The dataset in two formats allows us to be easily input into and trained with popular machine learning methods. The dataset also provides two sizes for all leukocyte images: 1) original size of 3264 × 3264, and 2) reduced size of 600 × 600 (mini size). The data in the former larger size can be used for error analysis, visual inspection or confirmation, and even further investigation. The data in the latter size can be used for model training, which helps to save a lot of CPU overhead time.
Model training: We build up a deep learning-based detector with leukocyte recognition capability using the training set. Training samples with ground truth labels are iteratively input into the training algorithm in mini-batches. The models output the predicted results, which are compared with the ground truth labels. The training process can be stopped when the training loss curve becomes smooth and the loss value is no longer decreasing in training. Finally, the internal parameters of the trained models are fixed and can be used in detecting leukocytes in new image samples from clinical practice. Inference: In the inference stage, the detectors are used in judging the new image samples it has never sees before. New image samples collected from the clinical practice are rescaled to the input size for the trained detectors. The output of inference from the detectors includes three data items, the recognized types of leukocytes, the confidence values, and the locations of the leukocytes in the image.
Evaluation: In the evaluation stage, the inferred results are evaluated with multiple metrics, and the performance of the model is also analyzed with different criteria. Evaluating metrics include mAP and mAR under the different values of IoU. Besides, we record executive performance, i.e., the size of the model, the inference speed in Frame Per Second (FPS), which helps to evaluate if the detectors are suitable or feasible to put into practice. As for accuracy, we emphasize the mAP because it is a popular and proven performance indicator in object detection. To further analyze the classification capability, we measure the average precision for each type of leukocyte.
Ensemble of predictions of deep learning-based models
In the domain machine learning, ensemble is a voting scheme that
takes account of predictions from different models. The way which
ensemble works is similar to the collective judgment by a medical
expert panel in diagnosing a complicated case. The advantage
of ensemble is that the final results are more stable for difficult
samples and potentially more accurate in quantitative evaluation.
On the other hand, however, it may cost more computational time
in inference. In this work, we integrate the ensemble scheme from
[30] into the post process stage and evaluate its results using the
leukocyte predictions from other deep learning-based models. The
ensemble [30] linearly combines the bounding boxes of leukocytes
with the corresponding confidences as the weights. Given a list
of overlapping predicted bounding boxes 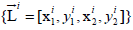 for
leukocytes and the corresponding confidence values {Ci} from
models (is generally less than or equal to, because of false negative
detection), the averaged bounding box
for
leukocytes and the corresponding confidence values {Ci} from
models (is generally less than or equal to, because of false negative
detection), the averaged bounding box 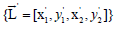 and the
updated confidence values {C '} are given as:
and the
updated confidence values {C '} are given as:
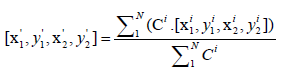
and
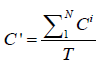
The former formula computes the averaged location by considering the confidences of predictions from different models: if a model is not so confident about its prediction, its vote is less powerful in influencing the final result. The latter formula suggests that the updated confidence values {C '} are averaged over the number of models rather than the number of predictions. In this way, if some models deem that the area has no leukocyte, which is like abstention, the updated confidence values {C '} will be lowered.
Implementation
The experiments are implemented on a regular workstation computer with Intel Core i5-8600 CPU, 16 GB RAM, and a Nvidia Titan Xp GPU with 12 GB graphic memory. The software environments are based on Ubuntu 18.04 OS, and training is carried out on PyTorch 1.7.0 (https://pytorch.org/) and mmdetection 2.6.0 codebases (https://github.com/open-mmlab/ mmdetection). The training epoch is set to 16, which is high enough for training convergence for the detectors. Setting this relatively excessive number of epochs is to ensure the models can approach its optimal state and avoid underfitting. The recorded training time for the models is around two hours.
That the models can be efficiently trained in such a short time is because of the use of pre-trained backbone networks and finetune technique. The finetune technique allows the model to shift its intelligence from recognizing generic objects to detecting leukocytes. The pre-trained weights are trained parameters from a deep learning-based model (Resnet-50 [31]) in classifying objects in ImageNet dataset or detecting objects in MS-COCO dataset. To finetune a model, we freeze the parameters in the low-level filters, which computes basic features of the image texture. On the other, high-level parameters, which reason the structural information, are gradually updated by backpropagation approach. In this way, it adjusts the high-level parameters in pre-trained weights to our leukocyte detection task.
When training the models as the key leukocyte detectors, we used Stochastic Gradient Descent (SGD) as the optimizer for the model parameter update. The key hype-parameters, i.e., the learning rate, momentum, and weight decay coefficient of SGD, are set to 0.01, 0.9 and 0.0001, respectively. Except for these settings, other detailed configurations for the architecture of detector are set defaults.
Data augmentation technique is employed in model training. Data augmentation is an online process that dynamically generates variants of training samples before feeding it into the model, following the sampling of the mini-batch of training data from the training set. Supplementary Table 1 shows a transformation list of data augmentation used in our implementation. The transforms are used as a composition with an occurrence probability for each transform.
| Sources | Types and Sample numbers | |||||
|---|---|---|---|---|---|---|
| Neutrophil images(cells) | Basophil images(cells) | Eosinophil images(cells) | Lymphocyte images(cells) | Monocyte images(cells) | Total images(cells) | |
| Training Set | 1214(2812) | 282(286) | 1006(1018) | 1232(1440) | 965(1031) | 4699(6587) |
| Test Set | 1015(1323) | 12(14) | 71(82) | 355(439) | 121(150) | 1574(2008) |
| Total | 2229(4135) | 294(300) | 1077(1100) | 1587(1879) | 1086(1181) | 6273(8595) |
Table 1: The number of images and leukocytes in the created dataset.
Establishment of a dataset with interference factors
The 111 wright-stained blood cell smears were collected from Tianjin Medical University Affiliated Medical Center (Tianjin Cancer Hospital and Tianjin Children's Hospital) and Rehabilitation Hospital of Hexi District, Tianjin. Five types of leukocytes from each smear were photographed by a Nikon DS-Ri2 Color Camera at 1000x original magnification for analysis. The five types of leukocytes include Neutrophil (NG), Basophil (BG), Eosinophil (EG), Lymphocyte (L), and Monocyte (M). Subsequentially, 6273 images in total were obtained, with nearly 2000 images containing multiple leukocytes, which is the first dataset suitable for leukocyte detection. These leukocytes images were divided into the training set and the test set with a ratio of 4:1.
Each sample in the dataset contains two items: A visual signal map in the form of the color image that may contain more than one type of leukocytes, and a manually labelled ground truth indicating the location(s) and type(s) of existing leukocyte(s). The ground truths are separately annotated by three experts with 18- year clinical experience using Labellmg toolkit software. During expert review and confirmation on the data samples, any cells with inconsistent type labels from different experts will be taken out. To avoid the problem of data imbalance, we have narrowed the statistical distribution gap among the five types of leukocytes as much as possible. Table 1 shows the composition of the five types of leukocytes in the training set and the test set. The processes of dataset formulation are also depicted in the corresponding part of the flowchart presented in Figure 1.
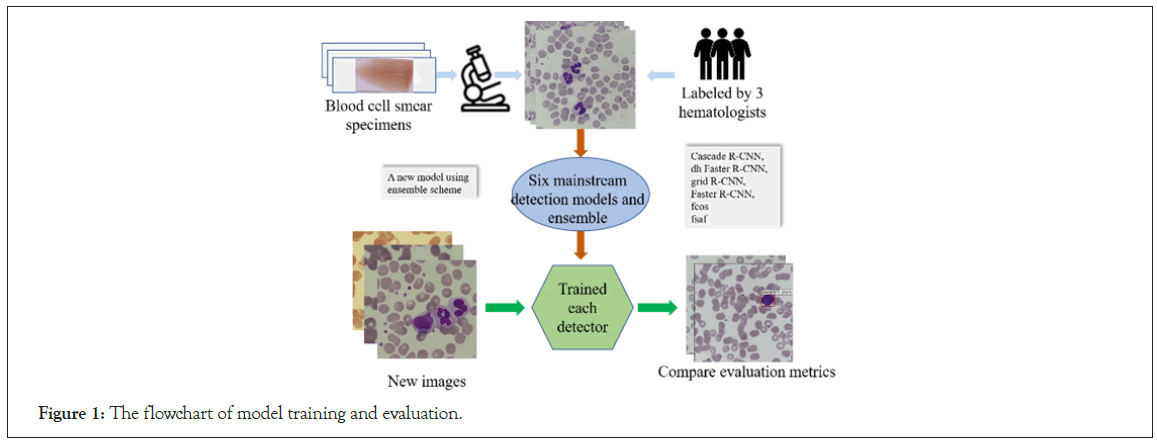
Figure 1: The flowchart of model training and evaluation.
Some “worse cases”, which are likely to appear in practical clinical scenarios, should be included in the prior knowledge (training set) for the AI model for assisting detection of leukocytes. The dataset we created contains nine factors that interfere with leukocytes detection, making the trained model generalize well. Some typical cases are shown in Figure 2. The statistics of the interference factors are manually collected and summarized. The specific numbers of these situations in the training set are shown in Supplementary Table 2. Meanwhile, these images contain multiple leukocytes, making the dataset more suitable for detection research. The statistical summaries of our dataset are presented in Supplementary Table 3.
| Model | mAP | mAP @IoU0.50 | mAP @IoU0.75 | mAR | AP-BG | AP-EG | AP-NG | AP-M | AP-L |
|---|---|---|---|---|---|---|---|---|---|
| FSAF | 0.742 | 0.874 | 0.860 | 0.866 | 0.618 | 0.684 | 0.870 | 0.756 | 0.785 |
| FCOS | 0.795 | 0.896 | 0.880 | 0.913 | 0.642 | 0.826 | 0.894 | 0.795 | 0.819 |
| Faster R-CNN | 0.815 | 0.940 | 0.919 | 0.879 | 0.738 | 0.847 | 0.893 | 0.785 | 0.814 |
| Grid R-CNN | 0.822 | 0.926 | 0.920 | 0.903 | 0.709 | 0.836 | 0.847 | 0.890 | 0.832 |
| DH Faster R-CNN | 0.848 | 0.951 | 0.939 | 0.910 | 0.753 | 0.892 | 0.908 | 0.842 | 0.843 |
| Ensemble | 0.853 | 0.940 | 0.933 | 0.922 | 0.752 | 0.898 | 0.925 | 0.843 | 0.848 |
| Cascade R-CNN | 0.856 | 0.948 | 0.938 | 0.909 | 0.781 | 0.905 | 0.916 | 0.838 | 0.841 |
Table 2: Performance in key evaluation criteria of six methods and ensemble scheme on test set.
Figure 2: Some exemplar samples are affected by the interference factors. a: Colour casts on blood cell smear images; b: Low illuminative intensity of blood cell smear images; c: Giant platelets; d: Incorrect imaging focal length of cell images; e: Images containing dyes or other impurities; f: Overlapping leukocytes; g: Degenerated leukocytes; h: Excessively high phosphate buffer solution (pH >6.8); i: Excessively low phosphate buffer solution (pH <6.4).
Performance comparison of the six models and ensemble result on test sets
We use the test set to evaluate the six well-trained detection algorithm models and draw the Precision-Recall (PR) curve of each model under different IOU thresholds, as shown in Figure 3. In Figure 3, it is not difficult to find that when the IOU threshold is lower than 0.8, all detectors/models output good results, and the AUCs are quite large. Among them, cascade RCNN and ensemble scheme are the TOP two among their competitors, FSAF is on par with other counterparts, and other models have average performance. When the IOU threshold is increased to 0.9, this means that the criteria for successful detection are more stringent. We can see that the divergence of the curves from each other increases. Now we can more easily distinguish the difference in performance. The advantages of cascade RCNN and ensemble scheme are more prominent. The corresponding specific quantitative results are shown in Table 2. For the test set, mAP@ IoU=0.50:0.95 of cascade RCNN is higher than the ensemble scheme (0.856>0.853), but its mAR@IoU=0.50:0.95 is 0.909, which is lower than that of the ensemble scheme (0.909<0.922). In addition, the Top 2 models have the best recognition performance on NG, and AP@IoU=0.50:0.95 is 0.916 and 0.925, respectively. The performance of BG recognition is low, AP@IoU=0.50:0.95 is 0.781 and 0.752, respectively. Table 2 also shows the corresponding detection performance indicators of other detection models.
Figure 3: The PR curve of each model at different IoU. Performance comparison of the six models and ensemble scheme on test sets. The x-axis represents the recall values, and the y-axis represents the precision. Note:
In this work, we also try to improve the performance by integrating an ensemble scheme as a post-process for the results. Although its mAP@IoU=0.50:0.95 is slightly lower than the Cascade RCNN among the tested models (0.853<0.856), its mAR@IoU=0.50:0.95 is the highest (0.922>0.909), which means that the ensemble scheme has the lowest rate of missed detection of leukocytes (Figure 3). That helps count leukocytes and prompts experts to verify the model to detect the wrong leukocytes. In addition, for the detection of leukocyte subtypes such as NG, M, and L, the ensemble scheme performs best, surpassing the cascade RCNN in the current evaluation models, which are shown in Table 2.
The performance comparison of detecting leukocytes with Cascade R-CNN and ensemble scheme
Detection of images is always challenging due to possible variance in staining, overlapping leukocytes, impurities, or even incomplete leukocytes. We deliberately considered these factors that easily affect the performance of the detection model on the dataset. In the test set, we focused on the detection effects of the Cascade R-CNN with the highest mAP values and the ensemble scheme. The accuracy of detecting leukocytes is 92.17% and 95.30% on the overly stained 447 images, respectively. Figure 4A shows the detection results of some images stained heavily. In addition, for the detection of overlapping leukocytes, the detection accuracy of Cascade R-CNN and the ensemble scheme are 65.79% and 94.74%, respectively. Figure 4B shows the detection performance of the model on leukocyte-dense scenes. From the results, the detection performance of Cascade R-CNN for dense scenes needs to be further improved. In the test set, there are 170 pictures containing impurities such as dye residues, cell debris, etc. The Cascade R-CNN and the ensemble scheme can better eliminate impurities when detecting leukocytes. The ratio of the impurities mistaken for leukocytes is 8.23% and 3.53%, respectively. Figure 4C shows the result of the model's detection of leukocytes containing impurities. In addition, the more surprising point is that the two models can accurately determine the type and location of incomplete leukocytes. The detection accuracy of incomplete leukocytes is 97.67% and 98.84%, respectively. Figure 4D shows the detection results of the model on incomplete leukocytes. Supplementary Table 4 shows the specific number of correct detections.
Figure 4: Some real examples of detecting leukocytes in different scenarios with Cascade R-CNN and the ensemble scheme. a: An example of excessively high phosphate buffer solution (pH >6.8) and small neutrophils. b: An example of overlapping leukocytes. c: An example of incomplete leukocytes. d: An example of impurities in the picture.
To further investigate the capabilities of the model, we analyze the classification performance of Cascade R-CNN and the ensemble scheme in a comparative way. The confusion matrix of Cascade R-CNN and the ensemble scheme is shown in Figure 5. In Figure 5A, we found that it was a bit difficult to identify eosinophils with Cascade R-CNN, and some neutrophils are mistaken for monocytes. Meanwhile, the accuracy of eosinophils or basophils was relatively lower than other types because that quite a part of eosinophils were misclassified into basophils. It is difficult for the ensemble scheme to identify basophils and eosinophils, and a small part of neutrophils are incorrectly identified as other types of leukocytes (Figure 5B).
Figure 5: The heat map of the confusion matrix for the Cascade R-CNN and the ensemble scheme’s performance in detecting five types of leukocytes on the test set.
In this study, we established a dataset with multi-leukocyte images, which took into account nine common interference factors in clinical application. Based on the research foundation and search results of the machine vision algorithm for detecting blood cells, we selected six detection models including Cascade R-CNN, dh Faster R-CNN, grid R-CNN, Faster R-CNN, FCOS, and FSAS for leukocyte detection. Cascade R-CNN has the best detection performance, mAP@IoU=0.50:0.95 is 0.856, and mAR@ IoU=0.50:0.95 is 0.909. Then we provide a powerful ensemble scheme. Without major modifications, the ensemble scheme can obtain high-performance indicators for leukocyte detection. mAP@IoU=0.50:0.95 is 0.853 and mAR@IoU=0.50:0.95 is 0.922. Through further in-depth analysis of the detection performance of Cascade R-CNN and the ensemble scheme, it is found that the ensemble scheme may be a better choice for the automated blood cell morphology system.
Datasets are the basis for solutions using data-driven artificial intelligence. The existing public leukocyte datasets [16-25] are either a small amount of data, or the images only contain a single leukocyte. These datasets are considered simple and cannot support the building up of an intelligent model for the complicated scenario in the clinic. To the best of our knowledge, this study created the first dataset with multi-leukocyte images that is close to the practical environment of clinical testing of peripheral blood smears. The dataset, as shown in Figure 2, takes account of nine interference factors that frequently happen in the clinical blood cell recognition process. The image is not limited to one leukocyte but includes multiple leukocytes, which is more suitable for the clinical environment. By considering these interference factors in the dataset and using the online data augmentation technique, the Cascade R-CNN and other models were trained on much wider data distributions, which increases generalization and alleviates the multi-center heterogeneity problem.
In the literature, research on using artificial intelligence to detect leukocytes is still limited. A report showed that mAP@IoU=0.5 used to detect normal leukocytes was 0.931 [27]. However, when we evaluate the performance of the detection model, we consider common clinical interference factors, which increase the difficulty of detection and are closer to the actual clinical detection environment. The mAP@IoU=0.5 of the Cascade R-CNN and ensemble schemes are 0.948 and 0.940 on the test set, which is both higher than 0.931 (Table 2). In addition, although the mAP@ IoU=0.50:0.95 of the ensemble scheme is slightly lower than that of Cascade RCNN (0.853<0.856), its mAR@IoU=0.50:0.95 is the highest (0.922>0.909), which means the integrated model has the lowest rate of missed detection of leukocytes (Figure 3). It helps to calculate leukocytes and prompts experts to verify the model to detect wrong leukocytes. In addition, for the detection of leukocyte subtypes such as NG, M, and L, the ensemble scheme performs best, surpassing the Cascade R-CNN model, as shown in Table 2.
The performance of the two models is further analyzed from the results of the challenging cases. Both Cascade R-CNN and the ensemble scheme are robust to significant pH changes, beyond the normal pH range (6.4, 6.8), and the models can accurately locate and identify poorly stained leukocytes, and the ensemble scheme performs better (95.30%>92.17%) (Figure 4A). Moreover, the detection ability of the ensemble scheme in the dense scenes is also higher than that of Cascade R-CNN, with a detection accuracy rate of 94.74%>65.79%, which makes the model has potential advantages in the detection of some leukemias (Figure 4B). In addition, in the clinical detection of leukocytes, common impurities including dye residue, broken red blood cells, dust, etc. frequently take place. These common impurities will affect the performance of the detection model. Both Cascade R-CNN and the ensemble scheme are robust to the interference of impurities. The latter performs better and has a low probability of misjudgment of impurities as leukocytes (3.53%<8.23%) (Figure 4C). It is worth noting that for the first time, this article found that both Cascade R-CNN and the ensemble scheme can detect the types of incomplete leukocytes with high accuracy [26,27,32]. The ensemble scheme is slightly better than Cascade R-CNN (98.84%>97.67%,), even for only covering 25–50% of the cells in a limited visible area, which means that the algorithm is superior to traditional image recognition algorithms (Figure 4D).
Regarding Cascade R-CNN and the ensemble scheme for the lowest detection results of basophils (Table 2 and Figure 5), research shows that in computer vision task more training examples can improve performance indicators, and the success of image classification tasks largely depends on the availability of labelled data [26]. A total of 8595 leukocytes were collected in our study. However, due to the different proportions of leukocyte types in the blood, the distribution of labels is not uniform. There are a total of 300 basophils in the training set and test set, while the numbers of other types of leukocytes are all >1000. Therefore, basophils showed the lowest mAP values in several models, ranging from 0.618 to 0.781. It is expected that with the expansion of the training set in the future and the increase of basophils, the performance of its detection will be significantly improved.
The error detection result map of the models may provide some correct suggestions to the clinical detection staff. As shown in Figure 6, we noticed that some detection errors are consistent among the top-three models, which all detected a monocyte as a lymphocyte. In the post review, it was noted that the leukocyte was considered to be an inaccurate marker by clinical experts. The leukocyte is an atypical lymphocyte. As we know, there are three subtypes of atypical lymphocytes: plasmacyte prototype (I), monocyte prototype (II), and prolymphocyte prototype (III), where the shape of subtype II is close to monocyte. It is difficult to accurately identify them in clinical practice, which needs to be combined with the overall blood smear of the subject and other test results to make a comprehensive judgment. Since only the photographed leukocyte images are provided to experts, it is more difficult to accurately identify the atypical lymphocytes, causing them to be mislabeled. Furthermore, that reflects the correct suggestions from the model to some extent. The deep network has learned a large number of image characteristics of leukocytes of different shapes, and the model summarizes the unique characteristics of distinguishing different types of them, which may improve the level of clinical detection of leukocytes. However, the model's ability of atypical lymphocyte detection needs to be strengthened in future work.
Figure 6: Qualitative true positive results yielded by different models. For the same leukocyte, Cascade R-CNN, Ensemble, and DH Faster R-CNN all detect it as lymphocyte, and experts labeled it as monocyte.
In summary, we believe that the ensemble scheme may be a better choice for automatic blood cell morphology systems. Using the ensemble scheme, the leukocyte detection process can reduce the dependence on experts, overcome the inherent limitations of clinicians' manual identification, such as the influence of subjectivity on identification, and improve the consistency of diagnosis. By taking the interference factors into consideration and training on collective knowledge from experts, the ensemble scheme “remembered” the accurate and wide-ranged prior information from the clinic. Therefore, the model is not only highly adaptive but also robust to daily clinical environments. Another advantage of the ensemble scheme is that its processing speed demonstrates a tremendous advantage over manual recognition and reduces workload.
In summary, our paper established a dataset with multi-leukocyte images, which took into account nine common interference factors in clinical application. On this basis, we evaluated 6 mainstream detection models and developed a new model to comprehensively evaluate their performance in terms of mAP@IoU=0.50:0.95, mAR@IoU=0.50:0.95, AP for each type of leukocyte, and robustness to different interference factors. We believe that the developed ensemble model can count leukocytes more accurately and prompt experts to detect wrong detections, and it is more robust. In addition, we found that the model's error detection result can provide clinical with some correct suggestions, which can help experts perform clinical testing.
This work was supported by the Science & Technology Development Fund of Tianjin Education Commission for Higher Education (2018KJ082 to S.Z.).
Xuguo Sun, Shijie Zhang, Cong Lin and Meiyu Li conceived and designed the study; Meiyu Li, Lei Li, Shuang Song and Peng Ge collected and curated the data; Shijie Zhang, Cong Lin, Hanshan Zhang, Xiaoxiang Liu, and Lu Lu designed and performed the experiments; Meiyu Li, Fang Zheng and Xuguo Sun analyzed the results; Meiyu Li and Cong Lin wrote the manuscript; Xuguo Sun, Shijie Zhang and Cong Lin revised manuscript. All authors contributed to the preparation of the manuscript.
Authors state no conflict of interest.
All blood smears involved in this study are historical samples. Since only blood smears from patients are photographed, the approval of the institutional review board is not required.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Li M, Li L, Song S, Ge P, Zhang H, Lu L, et al. (2022) A Deep Learning Model for Detection of Leukocytes under Various Interference Factors. J Clin Trials. 12:512.
Received: 13-Sep-2022, Manuscript No. JCTR-22-19182; Editor assigned: 15-Sep-2022, Pre QC No. JCTR-22-19182 (PQ); Reviewed: 29-Sep-2022, QC No. JCTR-22-19182; Revised: 06-Oct-2022, Manuscript No. JCTR-22-19182 (R); Accepted: 10-Oct-2022 Published: 13-Oct-2022 , DOI: 10.35248/2167-0870.22.12.512
Copyright: © 2022 Li M, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.