Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research - (2022)Volume 12, Issue 3
Production of phytohormones by plant-interacting Trichoderma species modifies the plant physiology to allow its colonization, and to promote plant growth and defense. Ethylene-dependent responses are triggered in plants inoculated with Trichoderma spp., but it is unclear whether such response is triggered by ethylene produced by Trichoderma itself. The rate-limiting step in ethylene biosynthesis is catalyzed by the enzyme 1-Aminocyclopropane- 1-Carboxylic Acid (ACC) Synthase (ACS). To gain insight into ethylene production in Trichoderma spp., the polypeptide sequences of six ACS-like proteins were identified in the endophytic T. harzianum. The identified ACS- like proteins have features of PLP-dependent enzymes and five of them conserve most of residues necessary to catalyze the biosynthesis of ACC. The predicted structure of ThACS-like1 illustrates a folding reminiscent of the apple enzyme MdACS1 and supports its ACS activity. An inspection into Trichoderma genomes confirmed that ACS- like1 is conserved in the unrelated species T. harzianum, T. atroviride, and T. reesei, although T. harzianum is enriched in ACS-like genes. These results demonstrate that T. harzianum, T. atroviride, and T. reesei might be endowed to synthesize ACC as a critical step to produce ethylene during their interaction with plants.
Trichoderma; ACC synthase; Ethylene; Plants
The ascomycete genus Trichoderma consists of species displaying a wide range of habitats and interactions with other organisms, whether fungi, plants, and animals [1]. Mycotrophy is a common trait among Trichoderma spp., which is characterized by the ability to recognize a fungal prey, directly invade hyphae, and utilize it as a nutrient source through the activity of its secreted hydrolytic enzymes [2]. Several soil-borne Trichoderma species can interact with plants to establish an endophytic growth. Trichoderma harzianum colonize and penetrate cucumber roots, these behavior shares similarities to the events of mycotrophy such as coiling and formation of appressoria-like structures [3]. Colonization of T. harzianum promotes plant growth by changing the phytohormone balance [4]. The genomes of Trichoderma contain putative genes involved in the biosynthesis of phytohormones such as auxins, cytokinins and gibberellins [5]. Since the production of auxins as well as ethylene has been evidenced, the in silico search for genes encoding phytohormone biosynthetic enzymes are a reliable approach to investigate the mechanisms underlying plant- Trichoderma interactions [5-7]. Trichoderma spp. produce volatile organic compounds that influence plant development [6], but ethylene has not been detected as part of the mixture of volatiles.
Ethylene is a gaseous phytohormone that controls several aspects of plant development such as fruit ripening, flower production, and mediates the response to biotic and abiotic stress [8]. Ethylene-dependent responses are triggered in plants inoculated with Trichoderma spp. [9,10]. This contributes to enhancing the response to pathogens; therefore, the protective effect of Trichoderma is not limited to mycotrophy, but we cannot rule out that the ethylene produced and perceived by the plant during interaction with Trichoderma mediates other aspects of plant development.
In plants, ethylene biosynthesis starts with the lysis of S-Adenosyl- L-Methionine (SAM) to 5’-methylthioadenosine (MTA) and 1-Aminocyclopropane-1-Carboxylic Acid (ACC), a reaction catalyzed by ACC Synthase (ACS), and the subsequent conversion of ACC to ethylene which is catalyzed by ACC Oxidase (ACO) [11]. ACC is also a signal molecule in plants that regulates cell elongation, stomatal development, and defense to pathogens [12]. ACS is a Pyridoxal 5’-phosphate (PLP)-dependent enzyme that belongs to the Fold Type I of the aspartate aminotransferase family [13]. The Aminotransferase Domain (ATD) harbors a conserved lysine that covalently binds to PLP, and the composition of neighbor residues of the catalytic site in cooperation with the PLP allows the catalysis of transferase and lyase reactions [14]. Although ethylene biosynthesis in fungi is a poorly explored topic, the production of ethylene by the supply of L-methionine in Trichoderma atroviride suggests that at least in this species, the ethylene biosynthesis takes place by a similar pathway to that which occurs in plants, with L-methionine as direct precursor of SAM [4]. A cDNA encoding an ACS enzyme has been cloned, and the recombinant protein showed ACS activity in vitro [15]. Nevertheless, the physiological consequence of ACC or ethylene in fungi is unknown as there are no characterized receptors of ACC or ethylene.
Here, it is presented the in silico search for potential ACS-like enzymes in the endophytic fungus T. harzianum (Harzianum clade) The analysis revealed 27 Fold Type I aminotransferases in T. harzianum, and six of these were classified as ACS-like enzymes. The prediction of the tertiary structure of ACS-like protein with higher conservation of catalytic residue supports the potential ACS activity. ACS-like enzymes are conserved in the divergent species T. atrovirde (Trichoderma clade) and T. reesei (Longibrachiatum clade) suggesting that biosynthesis of ACC and possibly ethylene production is a biochemical event that has taken place from the common ancestor to current species. These findings direct the future research towards to ethylene biosynthesis and function in Trichoderma species of different lineages and adapted to different habitats.
Protein alignment of phylogeny
The sequences of proteins containing the ATD (Interpro ID: IPR004839) of T. harzianum CBS 226.95 were retrieved from EnsemblFungi (https://fungi.ensembl.org/index.html) [16]. All the ID codes presented here were taken from EnsemblFungi. The sequences of previously characterized ACS enzymes and additional aminotransferase enzymes were obtained from the Universal Protein Resource database [17]. The plant ACS enzymes MdACS1(Uniprot ID: P37821) [18] and SlACS2 [13] were used as reference of actual ACS enzymes together with the Penicillium citrinum ACS (Uniprot ID: Q9P963) whose ACS activity is detectable [15]. The E. coli AAT (P00509) [19] and Giardia intestinalis AAT (A8B1V5) were used as ATDcontaining proteins unrelated to the ACS catalytic activity [20]. This whole set of polypeptide sequences was aligned by MUSCLE to later construct a maximum likelihood phylogenetic tree with 100 bootstrap replicates using PhyML 3.0 (http:// www.atgc-montpellier.fr/phyml) [21]. Proteins subclustered with the reference ACS enzymes were named ThACS-like proteins. MUSCLE alignment of ThACS-like proteins and plant ACS enzymes was repeated to identify conserved residues involved in catalysis. The ThACS-like proteins were named according to the similarity to plant ACS proteins, with ThACS-like1 being the most like plant ACS protein and ThACS-like6 showing the least similarity.
Modeling of ThACS-like1
The protein ThACS-like1 (EnsemblFungi ID: PTB53907) was selected to predict its tertiary structure in the Robetta Server (https://robetta.bakerlab.org) [22]. Six quite similar models were obtained, and of which was selected to be minimized, visualized and structurally compared against MdACS1 (3PIU) in the UCSF Chimera program [23].
Analysis of genes encoding ATD-containing proteins and conservation of ACS-like1 gene
The coding sequences of the whole family of ATD genes of T. harzianum CBS 226.95, T. atroviride IMI206040 and T. reesei QM6a were retrieved from EnsemblFungi. The presence of the IPR004839 domain was the criteria used to obtain all the ATD genes. Sequences were subsequently aligned by using ClustalW. A phylogenetic tree with 500 bootstrap replicates was constructed by the Bio Neighbor-joining algorithm [24]. To verify that ACS-like1 is part of a syntenic region, a sequence of 16.2 Kb that harbors the ACS-like1 gene of T. harzianum and an additional six annotated ORFs was searched in T. atroviride and T. reesei. Annotation was verified by the resubmission of the sequence in InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) [25]. This procedure was repeated to the whole set of genes encoded in the corresponding genomic fragment of T. atroviride and T. reesei. Once the gene annotation was confirmed, reciprocal BLASTX and TBLASTN among the corresponding polypeptide sequences of the three species were performed as criteria to identify the conservation of orthologs [26].
Identification of ACS-like proteins in Trichoderma harzianum
The ATD was identified in 27 proteins of T. harzianum. The sequence of this group of proteins was aligned together with the sequence of two previously characterized plant ACS enzymes and one known fungal ACS enzyme of P. citrinum. The sequences of EcAAT and GiAAT were included in this analysis as they have an ATD, but they catalyze an actual aminotransferase reaction unrelated to the characteristic lyase activity of ACS enzymes. The phylogeny presented in the Figure 1 shows that the ACS enzymes are clustered with six T. harzianum proteins with a high support value. The proteins of this subtree were named as ThACS-like. According to gene ontology, the subcellular localization and biological process of these proteins are unknown, but all of them have the assigned molecular functions of catalytic activity and PLP binding (data not shown).
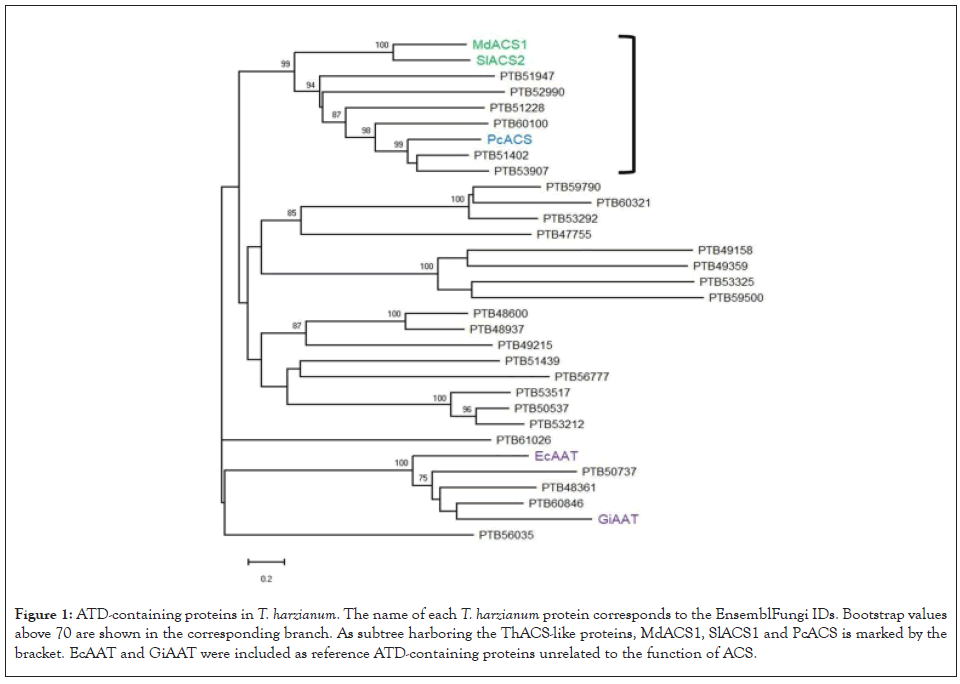
Figure 1: ATD-containing proteins in T. harzianum. The name of each T. harzianum protein corresponds to the EnsemblFungi IDs. Bootstrap values above 70 are shown in the corresponding branch. As subtree harboring the ThACS-like proteins, MdACS1, SlACS1 and PcACS is marked by the bracket. EcAAT and GiAAT were included as reference ATD-containing proteins unrelated to the function of ACS.
The rest of the ATD-containing proteins are grouped in independent subtrees with also a high support value, including the potential homologs of EcAAT/GiAAT. The crystal structure of MdACS1 and SlACS2 allowed the identification of critical catalytic residues involved in establishing hydrogen bonds with the cofactor PLP and the lysis of the substrate SAM [13,18]. Then, the conservation of such residues in the ACS-like proteins was assessed by aligning the plant ACS and ThACS-like sequences. The conserved catalytic residues in the ThACS-like proteins are shown in Table 1. PTB53907 (ThACS-like1) is the protein carrying more conserved residues that likely participate in the synthesis of ACC. The lysine involved in the binding of PLP is conserved in the rest of the ATD-containing proteins, although they lack the residues involved in the synthesis of ACC (data not shown).
Structure prediction of ThACS-like1 reveals a folding reminiscent to plant ACS
The predicted tridimensional model of the ThACS-like1 protein (Figure 2A) displays archetypic features of the ATD, and specifically matches the MdACS1 structure (Figure 2B). The overall structure of ThACS-like1 allows the projection of the residues mentioned in the Table 1, which might be involved in the catalysis (Figure 2A).
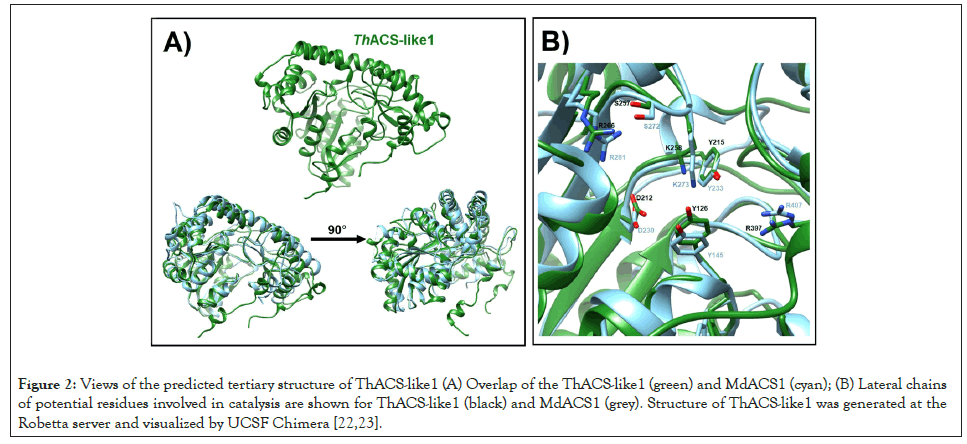
Figure 2:Views of the predicted tertiary structure of ThACS-like1 (A) Overlap of the ThACS-like1 (green) and MdACS1 (cyan); (B) Lateral chains of potential residues involved in catalysis are shown for ThACS-like1 (black) and MdACS1 (grey). Structure of ThACS-like1 was generated at the Robetta server and visualized by UCSF Chimera [22,23].
| Residue | Trichoderma harzianum proteins | |||||
|---|---|---|---|---|---|---|
| PTB53907 (ThACS-like1) | PTB51228 (ThACS-like2) | PTB51947 (ThACS-like3) | PTB60100 (ThACS-like4) | PTB51402 (ThACS-like5) | PTB52990 (ThACS-like6) | |
| A120 | - | + | - | - | - | - |
| T121 | + | + | - | - | - | - |
| Y144 | + | - | - | - | + | - |
| Y145 | + | + | + | + | + | - |
| N202 | + | + | + | + | + | + |
| D230 | + | + | + | + | + | + |
| Y233 | + | + | + | + | + | + |
| S270 | - | - | - | - | - | + |
| S272 | + | - | + | + | + | + |
| K273 | + | + | + | + | + | + |
| R281 | + | + | + | + | + | + |
| R407 | + | + | + | + | + | + |
Table 1: Residues in ACS-like proteins of T. harzianum potentially required to ACS activity. Residues were identified by aligning the polypeptide sequences with the MUSCLE algorithm. The corresponding residues to PLP-binding and catalysis in MdACS1 shown in the far-left column were used as reference [18].
Conservation of ACS-like genes in three Trichoderma species
The ATD-containing genes from T. harzianum, T. atroviride and T. reesei were retrieved from EnsemblFungi. These species belong to independent clades or sections within the Trichoderma genus [27]. Each species has a distinct habitat and traits, being T. harzianum a well-known endophyte [28,29]. T. atroviride is a soil-borne and river sediment fungus [27,30,31], that also interacts with roots [7], and T. reesei is a wood decomposer that produces hydrolytic enzymes for biotechnological applications [31]. The phylogenetic tree presented in the Figure 3 reveals conservation of homolog ACS-like genes in T. atroviride and T. reesei, including the ACS- like1 as a potential ortholog in the three species [32]. This family of genes is expanded in T. harzianum as six genes were identified in this species, in contrast to the three genes identified both in T. atroviride and T. reesei (Figure 3).
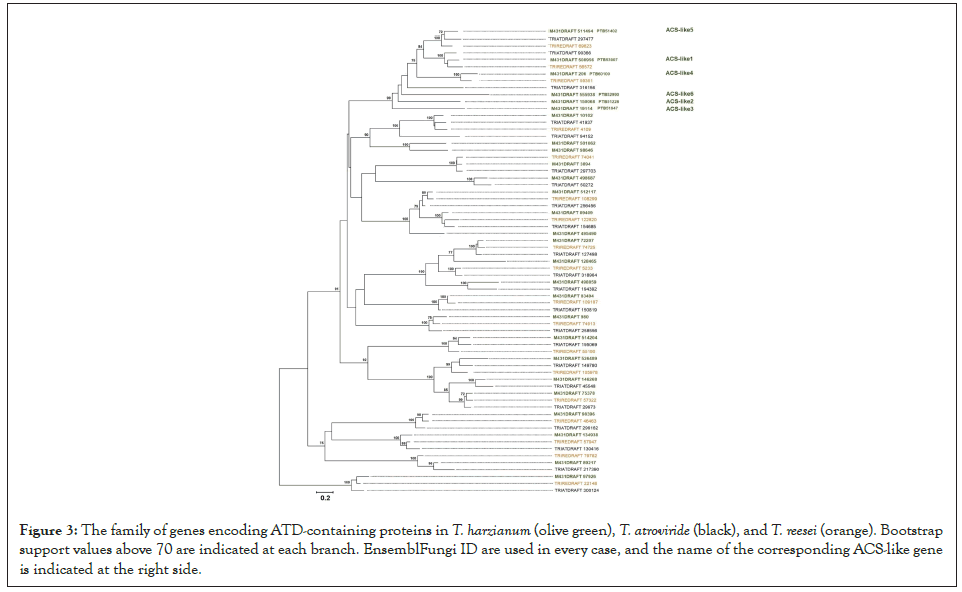
Figure 3: The family of genes encoding ATD-containing proteins in T. harzianum (olive green), T. atroviride (black), and T. reesei (orange). Bootstrap support values above 70 are indicated at each branch. EnsemblFungi ID are used in every case, and the name of the corresponding ACS-like gene is indicated at the right side.
A genomic region carrying ACS-like1 is conserved in 3 Trichoderma species
Gene annotation was performed for three genes upstream and three genes downstream of the ThACS-like1 gene. The diagram of Figure 4 shows the composition of this genomic fragment which encompasses 16.2 Kb. This process was repeated to the genomic fragments carrying ACS-like1 of T. atroviride and T. reesei. Reciprocal BLASTX and TBLASN among the respective surrounding genes confirmed that the best hit of each T. harzianum gene is the gene found in the genomic fragment of T. atroviride and T. reesei at the same position with respect to ACSlike1 (Figure 4). Genes encoding a Lysyl-tRNA synthetase, SAMdependent methyltransferase, and Syp26/Tex261 protein and located upstream ACS-like1, while an isocitrate dehydrogenase (IDH), a protein kinase and thioredoxin-domain containing protein are encoded downstream ACS-like1 in the three species. The ACS-like1 ortholog, and thus, the conserved region is lacking in the related fungus Escovopsis weberi (data not shown), suggesting this is genomic region is specific to Trichoderma spp.
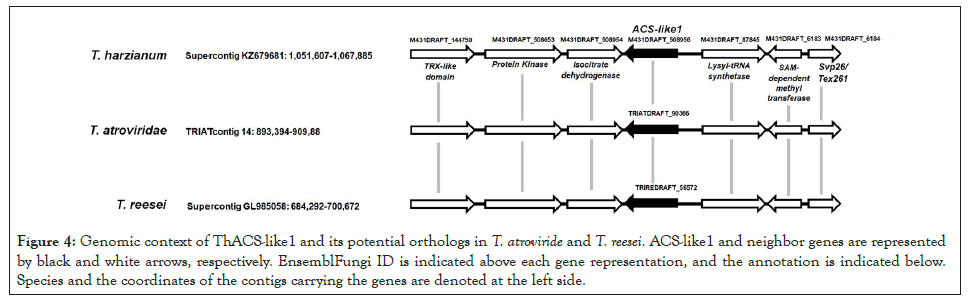
Figure 4: Genomic context of ThACS-like1 and its potential orthologs in T. atroviride and T. reesei. ACS-like1 and neighbor genes are represented by black and white arrows, respectively. EnsemblFungi ID is indicated above each gene representation, and the annotation is indicated below. Species and the coordinates of the contigs carrying the genes are denoted at the left side.
The enzymes containing the ATD mainly catalyze transferase, lyase and isomerase reactions. The distribution and variation of these enzymes within organisms might depend on the selection pressure that shapes the metabolic pathways necessary to survive in the habitat and respond to environmental cues. There are 27 ATD-containing proteins in T. harzianum; among them, six proteins share homology to plant and fungal ACS enzymes. The bootstrap values of the branches of the phylogenetic tree support this finding (Figure 1). The alignment of this subset of proteins together with plant ACS enzymes illustrates the overall similarity of the proteins (Figure 5). There are critical residues, including the lysine for PLP binding, residues involved in H bonds to set the orientation of PLP, and residues involved in catalysis. The alignment shows a conserved asparagine corresponding to the N202 of MdACS1 and the N209 of SlACS2 (Figure 5). The amino group of N209 of SlACS2 forms hydrogens bonds with the cofactor PLP [13]. Y240 in SlACS2 and Y233 in MdACS1 set hydrogen bonds with PLP [13]. The equivalent residue in ThACSlike1 is Y215 (Figure 5). Plant proteins have two serine residues at the N terminus of the catalytic lysine, the serine closest to the catalytic lysine is conserved in ThACS-like proteins, and it is necessary to set hydrogen bonds to the phosphate moiety of PLP and interacts with the ACS inhibitor L-vinylglycine [13,14]. A tandem of tyrosine residues, Y151 and Y152 of SlACS2 are required to catalyze the conversion of SAM to ACC and MTA [13]. The equivalent residues in MdACS1 are Y144 and Y145 (Figure 5).
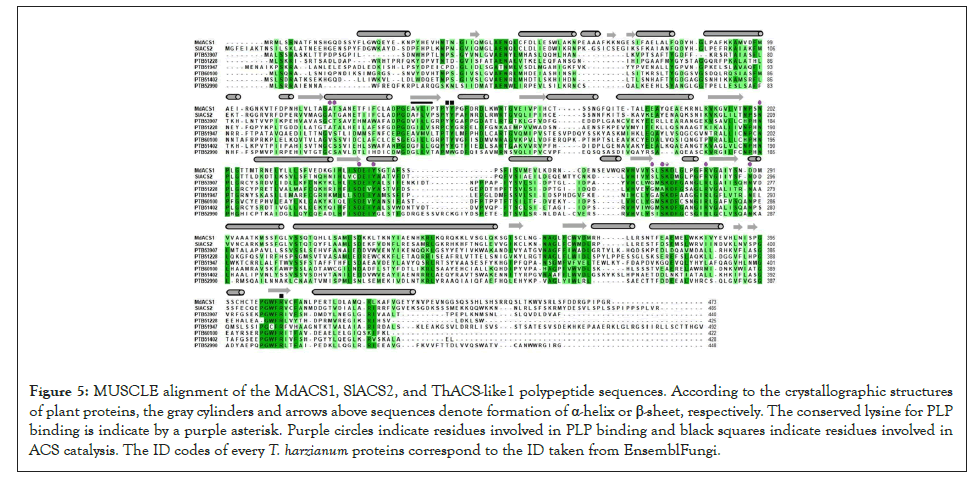
Figure 5: MUSCLE alignment of the MdACS1, SlACS2, and ThACS-like1 polypeptide sequences. According to the crystallographic structures of plant proteins, the gray cylinders and arrows above sequences denote formation of α-helix or β-sheet, respectively. The conserved lysine for PLP binding is indicate by a purple asterisk. Purple circles indicate residues involved in PLP binding and black squares indicate residues involved in ACS catalysis. The ID codes of every T. harzianum proteins correspond to the ID taken from EnsemblFungi.
Substitution of both residues, especially Y152, reduces the catalysis [33]. Both residues are conserved in ThACS-like1 and ThACS-like5 (Table 1). ThACS-like2, ThACS-like3 and ThACSlike4 only conserve the second tyrosine that has a more prominent role in the catalysis. Although ThACS-like6 shares homology with actual ACS enzymes, it lacks such catalytic tyrosines, thus, its ACS activity is unlikely. In accordance with the results of the results, the modeling of ThACS-like1 reveals features of ATD such as the β-sheets covered with helices [33] (Figure 2A). Overlapping of ThACS-like1 and MdACS1 illustrates that ThACS-like1 maintains the position of catalytic residues, including the abovementioned catalytic tyrosine Y126 that corresponds to the Y245 in MdACS1 (Figure 2B). Mutation R407K in MdACS1 increases the Km value of SAM and it is conserved in SlACS2 [13]. This residue is also conserved in the six ThACS-like proteins (Figure 5), including the R397 of ThACS-like1 that is part of the predicted catalytic site (Figure 2B). Thus, this predicted model exemplifies that ThACS-like1 displays a tertiary structure that enables its ACS activity. However, the conservation of residues of the ACS-like proteins is not conclusive evidence of ACS activity. The ATD-containing proteins of yeast ScAlt1 and ScAlt2 are annotated as alanine transaminases that share high identity and conservation of catalytic residues. However, ScAlt2 has a more open conformation that impedes the alanine transaminase activity [34]. Therefore, the catalytic activity of the ATD depends on the overall tertiary structure, but out of the full set of ATD-containing proteins of T. harzianum, the findings presented here narrowed down a concise group of enzymes reminiscent to the plant ACS enzymes in this endophyte that might produce ACC or ethylene as part of its strategy to interact with plants. A phylogenetic tree of the coding sequences of ATD-containing proteins from three Trichoderma species was constructed to answer how conserved and distributed the ACS-like genes are. To avoid the bias, the species T. harzianum, T. atroviride and T. reesei were selected as they belong to independent clades from the Trichoderma genus. Particularly, T harzianum has higher metabolic diversity [35], this species is a known endophytic fungus that might require ACC or ethylene to cope with living plant cells at the apoplast. This phylogenetic tree reveals that the six genes encoding ACS-like in T. harzianum are clustered with three genes of T. atroviride and T. reesei (Figure 3). The genes ACS-like1 and ACS-like5 are conserved in the three species and are potential orthologs, ACS-like4 is conserved in T. harzianum and T. reesei, and ACS-like2, ACS-like3 and ACS-like6 are unique paralogs of T. harzianum. T. atroviride that interacts with Arabidopsis roots and produces ethylene in vitro by exogenous addition of SAM [4] has one specific ACS-like gene (Figure 3). T. reesei lacks unique genes within this subtree. T. atroviride interacts with roots of Arabidopsis. The expanded number of ACSlike genes in T. harzianum suggests their role to produce ACC or ethylene as a diffusible factor during plant colonization, but the contribution of these proteins to the formation of ACC or ethylene is a future task. Therefore, we have a scenario where the ACS-like1 gene could be derived from an aminotransferase encoded in the genome of the ancestor of the Trichoderma species presented here. Ethylene and ACC itself play roles in plantmicrobe interactions [8,12], and their perception by the plant might affect the induction of defense related genes in systemic tissue [7,10]. The signaling crosstalk between phytohormones is necessary to plant adaptation to frequent abiotic and biotic stresses, and to exploit nutrient sources to maintain the growth [36]. Therefore, the role of ACC or ethylene might not be limited to local and systemic defense; it could be part of a more complex framework that regulates development and nutrient acquisition in concert with the interaction with microbes, whether beneficial or pathogenic. The fungal pathogen Verticillium dahliae which produces ethylene carries an ACC deaminase (ACCd) that catalyzes the conversion of ACC to α-ketobutyrate and ammonia [37]. Overexpression of ACCd enhances virulence while its disruption decreases symptom development. In accordance with the overexpression, the levels of ACC decreased when compared to the wild-type strain. The ACCd overexpression strain displayed enhanced virulence and increased in planta fungal growth when compared to the wild-type strain [38], suggesting that virulence is favored by lowering the levels of ACC whether produced by the fungus or by the plant. A putative gene encoding a ACCd was identified in T. harzianum (XP_024771635.1), T. atroviride (XP_013945914.1) and T. reesei (XP_006967764.1), and putative ACO genes were also identified in the three Trichoderma spp (data not shown). Therefore, a potential interplay between ACS-like and ACCd might regulate the levels of ACC to mediate certain responses during the interaction with plants.
An expansion in the number of ACS-like sequences in the T. harzianum by gene duplication might be necessary to mediate the accumulation of ACC or ethylene in the soil, rhizosphere or during different stages of its endophytic growth as a mechanism to cope with the plants, whether as a direct phytohormone or as a precursor of ethylene. Future work should address whether these enzymes play redundant roles or are involved in specific stages of plant colonization, and whether the endophytic growth occurs before root penetration. Genomic fragments containing the conserved gene ACS-like1 and six flanking genes were analyzed to determine whether ACS-like1 is encoded into a syntenic region in the three species. The annotation of such surrounded genes was confirmed; most encode for enzymes or carry a putative catalytic domain. Figure 4 illustrates the composition of the genomic fragment that is conserved in the three species. The coordinates of each contig are indicated, and they are available in EnsemblFungi. Large syntenic regions and the conservation of gene order and orientation are rarely found in fungal genomes [38]. Due to intra-chromosomal recombination and activity of transposable elements, the evolutionarily conserved genes of fungi do not maintain co-linearity; they rather present mesosynteny, defined as the conservation of genes without maintaining order and orientation [39]. However, fungi possess microsyntenic regions harboring from two to about ten genes that retain order and orientation [38]. Due to their size and potential biological relevance, those regions were unaltered during evolution. Genes of the microsyntenic regions are often involved in a common molecular process, and they form a metabolic gene cluster when the genes encode for enzymes of metabolic pathways of specialized or adaptive functions [40]. The 16.2 Kb regions containing the ACS-like1gene has features of the microsyntenic region since it is conserved in Trichoderma spp. and preserves the order and orientation of the surrounding genes, but the region is unlikely to be a metabolic gene cluster. Three conserved genes are located upstream ACS-like1 genes. Two of the genes are annotated as LysyltRNA synthase, which catalyzes the formation of Lysyl-rRNA that carries Lysine residues to the ribosomes, a Syp26/Tex261protein, which is a yeast protein involved in the transport of vesicles from the endoplasmic reticulum to Golgi apparatus [41] (Figure 4).
A search in InterPro [25] confirmed both carry characteristic domains of Lysyl-tRNA synthase and Syp26/Tex261, respectively. Due to their potential function, those genes could not be linked to the biosynthesis of ACC or ethylene. The other upstream gene encodes for a SAM-dependent methyltransferase (Figure 4). This protein also carries an 18S rRNA [guanine(1575-N(7)] methytransferase Bud23-like C-terminal domain, which suggest it is involved in the methylation of rRNA [42]. Although the potential function of this methyltransferase is not directly related to the activity of ACC synthase, its activity might affect the availability of SAM to ACC biosynthesis. The three conserved genes located downstream ACS-like1 encode an IDH, a protein kinase and a thioredoxin (Figure 4). The IDH catalyzes the decarboxylation of isocitrate to -ketoglutarate as part of the reactions of the citric acid cycle [43]. The polypeptide also carries a transit peptide to mitochondrion. Disruption of the IDH2 gene in the yeast Saccharomyces cerevisiae reduced the growth rate and disabled the yeast to utilize glycerol or lactate as carbon sources [43]. Hence, there is not clear link between the synthesis of ethylene, ACS activity or ATD-containing proteins and the potential function of the genes at the nearby of ACS-like1. Species of the Harzianum, Longibrachiatum, and Trichoderma clades share 7000 orthologs, and the formation of such clades occurred 20 to 30 million years ago, during the Oligocene [27]. The consistency in the position of the genes presented here suggests they have been part of a microsyntenic region of ortholog genes formed early in the diversification of the clades mentioned above. A potential selection pressure could keep these genes clustered, even if no functional relationship between such genes is found. In future work, the biological significance of ACS-like1 should be demonstrated and compared in species of different clades.
The identification of ACS-like proteins in T. harzianum and the structural prediction of ACS-like1 are consistent with the notion that T. harzianum produces ACC or ethylene. Although the activity of these enzymes must be experimentally validated in future work, the findings of this report support the metabolic richness of T. harzianum and its coupling into events that occur during the interaction with plants. The in silico identification of ACS-like genes in T. atroviride and T. reesei suggests this reaction is evolutionary conserved, and it might be crucial to the molecular communication with plants.
The author thanks to Tania Reyes for bioinformatic assistance. The author is supported by the Cátedras CONACYT Research Program, Grant No. 538
[Crossref], [Google Scholar]
Citation: Maldonado-Bonilla LD (2022) A Group of Aminotransferase Domain-Containing Proteins in Trichoderma harzianum Resembles Enzymes Catalyzing Acc Synthesis. Fungal Genom Biol. 12:187.
Received: 22-Apr-2022, Manuscript No. FGB-22-17114; Editor assigned: 27-Apr-2022, Pre QC No. FGB-22-17114 (PQ); Reviewed: 11-May-2022, QC No. FGB-22-17114; Revised: 17-May-2022, Manuscript No. FGB-22-17114 (R); Published: 24-May-2022 , DOI: 10.35841/2165-8056.22.12.187
Copyright: © 2022 Maldonado-Bonilla LD. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.