Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research - (2021)Volume 10, Issue 4
Small Cell Lung Cancer (SCLC) is a multistage, aggressive, metastatic form of lung cancer that accounts for about 13% of all lung cancer cases, with a 5-year survival rate of less than 6% mainly due to its metastatic component. The focus of metastatic SCLC has been focused on inhibiting the communication between SCLC circulating tumor cells and Cell Adhesion Molecules (CAMs) on endothelial cells, which cancer cells utilize to facilitate transmigration into a secondary tissue site after detaching from the primary tumor. It is well known that multiple CAMs are involved in SCLC transmigration; however, due to the different structures of CAM superfamilies involved in communication, efficient inhibitors to multiple CAMs have not been recognized. This study presents an in silico, molecular modeling approach to find potent inhibitors to multiple cell adhesion molecules involved in SCLC metastasis, including ICAM-1, VCAM-1, E-selectin, and P-selectin. From the ZINC15 database, 13 ligands of similar structure to Manassantin A and Casearinol A, two potent inhibitors of E-selectin and ICAM-1, were obtained and screened according to Lipinski’s Rule of 5. After preprocessing the proteins and ligands for docking, the ligands were blind docked to each CAM through AutoDock 4.0, and the binding affinities for each interaction were recorded. Docking results revealed four ligands (Ligands 8, 9, 10, 12) that recorded binding affinities significantly higher than that of Manassantin A and Casearinol A for E-selectin, P-selectin, and VCAM-1, while Ligand 11 (ZINC000146747094) recorded greater binding affinities than both controls across all four cell adhesion molecule groups. This study identified potent inhibitors against multiple cell adhesion molecules involved in SCLC tumor progression. Future experiments should test these inhibitors in vitro on SCLC cell lines to verify binding affinities.
Small cell lung cancer; E-selectin; P-selectin; Cell adhesion molecules
Small Cell Lung Cancer (SCLC) is a multistage, aggressive, metastatic form of cancer that occurs in 13% of all lung cancer patients and is directly correlated to smoking (Cleveland Clinic). SCLC has a very poor 5-year survival rate of less than 6%, ranking it among the deadliest forms of cancer. Furthermore, the main reason for this poor prognosis is due to its metastatic component, which describes the spread of SCLC cells to create a secondary tumor site. The progression of SCLC can be divided into two main stages: the limited stage and extensive stage [1]. During the extensive stage, the cancer metastasizes to both lungs, lymph nodes on the other side of the chest, or travels throughout the body to the bone marrow, liver, or brain. More than ⅔ of cases are caught during the extensive stage, when the cancer becomes inoperable and common treatments are no longer viable [2].
Metastasis of SCLC is the aggressive and rapid spreading of the tumor to essential organs and is a multistep process that is responsible for the deaths of 90% of lung cancer patients [3]. Furthermore, excising the tumor at secondary tissue sites would mean extremely risky surgery, possibly interfering with major functions of the body and leading to death. Metastasis of SCLC begins with the formation of a primary malignant tumor, usually attached to the lungs or surrounding lymph nodes [4]. Once the primary tumor grows to a large enough size, the tumor cells secrete extracellular matrix proteins that allow them to detach from the primary malignant site, allowing them to enter the bloodstream in a process called intravasation. Following intravasation, SCLC travels at high velocities through the bloodstream as circulating tumor cells and begin slowing down at a secondary attachment site. Once the cancer cells have stopped, they communicate with the vascular endothelium, a lining of endothelial cells on blood vessels that act as a gateway to organs and tissues. Here, they interact with cell adhesion molecules, a lining of glycoproteins on endothelial cells that normally aid with leukocyte immune response by signaling for inflammation of a tissue site (Springer). However, SCLC essentially tricks these cell adhesion molecules by appearing as leukocytes, allowing them to attach to endothelial cells through receptor-mediated attachment (Figure 1). After attachment, the tumor cells break through the endothelial wall into a secondary tumor site in a process called extravasation, attaching to nearby organs, and forming a secondary tumor site, completing metastatic progression. As a result, the tumor cells can inhibit an organ’s function by multiplying and taking away nutrients from healthy cells, ultimately leading to organ failure (Figure 2).
Figure 1: Preprocessed structure of E-selectin.
Figure 2: Preprocessed structure of P-selectin.
One of the main focuses of metastatic cancer treatment is focused on preventing the interaction between cancer cells and cell adhesion molecules, ultimately preventing extravasation into a secondary tumor site. Smith and Springer each studied cell-surface adhesion molecules on endothelial cells and found that leukocyte adhesion molecules interact with cancer cells and allow them to bind onto endothelial cells [4,5]. Springer further classified these cell adhesion molecules into categories based on their structures: integrins, selectins, immunoglobulins, and cadherins [6]. Taken together, they pinpointed a few cell adhesion molecules on endothelial cells that are involved in the interaction with tumor cells, including E-selectin, P-selectin, L-selectin, ICAM, VCAM, β integrin, P-Cadherin, and N-Cadherin. Heidemann et al. used gene knockout models to rid SCLC-inflicted mice of certain combinations of selectins, including E-selectin and P-selectin; although this significantly reduced adhesion of SCLC to the vascular endothelium, it was determined that multiple cell adhesion molecules are simultaneously utilized by SCLC tumor cells in order to extravasate into a secondary site [4]. Additionally, Kobayashi et al. specified ICAM-1, VCAM-1, E-selectin, and P-selectin as most involved in cancer metastasis, common across many metastatic cancers such as lung cancer and breast cancer, making them targets for future inhibition [7]. Ultimately, attempts to prevent metastasis of SCLC have been focused on inhibiting cell adhesion molecule receptors expressed on vascular endothelial cells; however, a potent inhibitor of multiple cell adhesion molecules simultaneously has not been found.
Molecular docking and virtual screening are important parts of the drug discovery process and have low-cost implications with high effectiveness. In essence, docking is a method that tests the effectiveness of molecules that bind to each other and predicts the preferred binding orientation of one molecule to another to create a stable complex Meng et al. [8]. Molecular docking uses machine- learning trained programs to dock a library of molecules on a protein, to predict whether the molecules are effective enough to inhibit the protein’s function. Forli et al. outlined specific molecular docking programs with high efficacy including AutoDock 4.0, the newest version of AutoDock available for public use [9]. Moreover, they describe the specific steps needed to be carried out prior to docking, including retrieving ligands and proteins, preparing each for docking, and utilizing a scoring function to calculate binding affinities. Ultimately, molecular docking is a cost-effective method to pinpoint inhibitors of chosen proteins and is essential to drug discovery.
To specify ligands for molecular docking testing, current potent inhibitors need to first be identified. Takamatsu identified many naturally occurring cell adhesion molecules inhibitors, the most potent of which are Manassantin A, isolated from Saururus chinensis, and Casearinol A, isolated from Casearia guianensis. Both Manassantin A and Caseariniol A effectively inhibited E-selectin and ICAM-1 on HUVECSs in a dose-dependent manner, without cytotoxicity and with very high potency [10]. Additionally, both Manassantin A and Casearinol A satisfied Lipinski’s Rule of 5, a standard employed by pharmaceutical companies to determine to determine if a drug has chemical properties that would make it orally active in humans, evaluating molecular weight, hydrogen- bond donors and acceptors, and lipophilicity (LogP) (Drugbank). Moreover, ligands of similar structure to Manassantin A and Casearinol A could potentially be considered as targets for cell adhesion molecule inhibitors [11].
Collecting information from previous research, a novel approach to studying SCLC metastasis would be to identify potent ligand inhibitors to cell adhesion molecules involved in lung cancer metastasis. In order to locate a specific cell adhesion molecule inhibitor, a molecular docking procedure can be performed, and ligands that are analogs to Manassantin A and Casearinol A, two naturally-derived, potent cell adhesion molecule inhibitors, can be screened as per Lipinski’s Rule of 5. It was hypothesized that if a protein-ligand blind docking procedure is performed and ligands that are analogs to Manassantin A and Casearinol A are screened, a potent inhibitor for each target cell adhesion molecule will be found and will satisfy Lipinski’s Rule of 5. Furthermore, ligands and proteins will be retrieved from online protein/ligand data banks, screened using Lipinski’s Rule of 5, converted to the appropriate file format, preprocessed for docking, and docked using the AutoDock 4.0 molecular docking simulation. The independent variable in this study are the various analogs of Manassantin A and Casearinol A chosen for docking and the dependent variable is the binding affinity of the ligands to each cell adhesion molecule. The constants include population size during docking, algorithm used, number of docking trials per ligand-protein simulation, and method of testing. The two controls in this study are Manassantin A and Casearinol A, two potent cell adhesion molecule inhibitors [12].
Prior to the start of the experiment, all appropriate software and programs were downloaded for all steps of the experiment. The application/database and their uses are shown in the table below, and further described in the procedures (Table 1) [13-15].
| RCSB Protein Data Bank (PDB) | Downloading protein structures and finding heteroatoms |
|---|---|
| ZINC15 database | Downloading ligands and both controls |
| Open Babel GUI | Converting ligands from .pdb to .pdbqt format |
| AutoDock tools 1.5.6 | Preprocessing proteins, visualizing proteins and ligands |
| AutoDock 4.0 | Docking ligands to protein, calculating binding affinities of each interaction |
| PyMOL visualization software | Specifying binding site residues on the protein, visualizing the 3-dimenstional protein-ligand attachment |
Table 1: Various applications/databases and their respective uses.
1. Structures for each cell adhesion molecule protein were downloaded from RCSB Protein Data Bank.
a) Each of the four cell adhesion molecule proteins were found on PDB, downloaded into .pdb file format, and saved into a file folder.
b) Protein PDB IDs:
• E-selectin PDB: 4CSY
• ICAM-1 PDB: 1IC1
• P-selectin PDB: 1G1R
• VCAM-1 PDB: 1VSC
2. Ligands were selected from the ZINC15 substance database, using Manassantin A and Casearinol A as controls.
a) 7 natural analogs of Manassantin A and 6 natural analogs of Casearinol A were randomly selected from ZINC15 “Substances” tab and downloaded individually as .pdb files.
b) Manassantin A and Casearinol A were downloaded individually as .pdb files.
c) Each ligand was saved individually to the file folder with the protein .pdb files.
3. Ligands were inputted into a Lipinski’s Rule of 5 Calculator and screened for drug-like, orally active characteristics.
4. Proteins were preprocessed using Auto Dock Tools 1.5.6 and downloaded as .pdbqt files.
a) From each protein, heteroatoms and water molecules were removed, atoms were repaired, polar hydrogens were added, and Kollman charges were added to each protein.
i. To find which heteroatoms to remove, the “WebGL” view of each protein on the PDB website was used, and the dropdown menu with pre-attached ligands were removed in Auto Dock Tools.
b) Proteins were gridded using the “Grid Box” feature, and the (x, y, z) coordinates were saved as text files into the file folder.
5. Ligands were preprocessed with Open Babel GUI and downloaded as .pdbqt files.
a) Each ligand .pdb file, including controls, was individually inputted into Open Babel GUI, converted from .pdb to .pdbqt files on the interface, and saved into the file folder.
6. Blind-docking procedure was performed on AutoDock Tools, using AutoDock 4.0 software.
a) Each of the ligands, starting with the controls, was inputted into AutoDock Tools along with one of the four proteins.
b) For each ligand-protein combination, the “AutoGrid” and “AutoDock” procedures were carried out, using a population size of 300, 50 runs, and a Lamarckian genetic algorithm for each ligand- protein docking.
c) The binding affinities for each of the 50 runs were compiled in a .dlg text file, and the top binding affinity for each ligand-protein interaction was recorded in a data table.
7. The ligand with the highest ranked binding affinity was inputted into PyMOL Visualization software
a) The .pdbqt protein file and the ligand_out file produced by AutoDock 4.0 were opened in the PyMOL Visualization software
b) The 3-dimensional view of the protein-ligand interaction was visualized on PyMOL, using the “Surface View” option.
c) The polar residues, which were amino acids attached to hydrogen bonds (indicated by a dotted yellow line) on the protein, were selected using the PyMOL select tool.
d) The polar residues most involved with ligand binding were recorded for each protein and the protein-ligand visualization was downloaded.
Data: Prior to the docking procedure, the proteins were
preprocessed in AutoDock Tools 1.5.6. The four preprocessed proteins in the AutoDock Tools interface are shown (Figures 1-4). 
Figure 3: Preprocessed structure of VCAM-1.
Figure 4: Preprocessed structure of ICAM-1.
Following each ligand-protein docking simulation, the highest ten binding affinities of all 50 trials (per ligand-protein combination) were recorded in the .dlg file produced by AutoDock 4.0. The strongest of these ten values, for each ligand-protein combination (including the controls), was then recorded in the data table shown (Table 2).| Cell adhesion molecule | Manassan tin A bindinging affinity | Cassearinol A bindinging affinity | Ligand 1 A bindinging affinity | Ligand 2 A bindinging affinity | Ligand 3 A bindinging affinity | Ligand 4 A bindinging affinity | Ligand 5 A bindinging affinity | Ligand 6 A bindinging affinity | Ligand 7 A bindinging affinity | Ligand 8 A bindinging affinity | Ligand 9 A bindinging affinity | Ligand 10 A bindinging affinity | Ligand 11 A bindinging affinity | Ligand 12 A bindinging affinity | Ligand 13 A bindinging affinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-selection | -7.16 | -7.06 | -4.34 | -2.26 | -3.67 | -5.73 | -5.11 | -5.03 | -6 | -7.16 | -7.68 | -7.83 | -8.89 | -7.98 | -5.87 |
| P-soloction | -2.66 | -5.55 | -1.93 | -3 | -1.47 | -2.39 | -2.92 | -2.55 | -2.52 | -6.46 | -7.22 | -6.59 | -7.21 | -7.08 | -4.31 |
| VCAM-1 | -4.94 | -5.6 | 32.25 | 48.57 | 34.91 | 47.87 | 26.55 | 11.18 | 11.18 | -5.62 | -6.06 | -6.45 | -7.61 | -8.86 | -5.25 |
| ICAM-1 | -3.59 | -1.94 | 2.17 | -0.35 | 1.43 | 1.89 | 1.27 | 0.72 | 0.72 | -2.33 | -2.43 | -2.21 | -3.67 | -2.81 | -0.81 |
Table 2: Highest recorded binding affinities (kcal/mol) of 13 different analogs of Manassantin A and Casearinol A to cell adhesion molecules E-selectin, P-selectin, VCAM-1, and ICAM-1.
After recording the highest binding affinities of each ligand-protein docking, the data was processed to draw conclusions on which ligands were the strongest, both compared to the other ligands and the two controls. To do so, the ligands, along with the controls, were ranked from strongest to weakest binding affnity, shown (Table 3). Next, the ligand structures of the five highest ranked ligands (Ligands 8-12) were retrieved from ZINC15 and compared to Manassantin A and Casearinol A, to notice similarities in structure. The ligand structures are shown (Figure 5).
| Ligand | Binding affinity Rank: E- soloction | Binding affinity rank: P- soloction | Binding affinity rank : VCAM-1 | Binding affinity rank : ICAM-1 |
|---|---|---|---|---|
| Manassantin A | 5 (tie) | 10 | 8 | 2 |
| Casearinol A | 6 | 6 | 6 | 7 |
| Ligand 1 | 12 | 14 | 12 | 14 |
| Ligand 2 | 14 | 8 | 15 | 9 |
| Ligand 3 | 13 | 15 | 13 | 14 |
| Ligand 4 | 9 | 13 | 14 | 15 |
| Ligand 5 | 10 | 9 | 11 | 12 |
| Ligand 6 | 11 | 11 | 10 | 11 |
| Ligand 7 | 7 | 12 | 9 | 10 |
| Ligand 8 | 5 (tie) | 5 | 5 | 5 |
| Ligand 9 | 4 | 1 | 4 | 4 |
| Ligand 10 | 3 | 4 | 3 | 6 |
| Ligand 11 | 1 | 2 | 2 | 1 |
| Ligand 12 | 2 | 3 | 1 | 3 |
| Ligand 13 | 8 | 7 | 7 | 8 |
Table 3: Binding affinity rank (strongest to weakest) of ligands to each cell adhesion molecule.
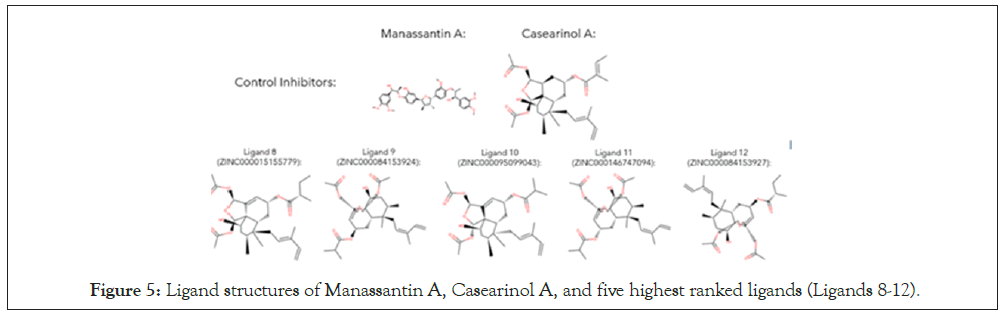
Figure 5: Ligand structures of Manassantin A, Casearinol A, and five highest ranked ligands (Ligands 8-12).
The Lipinski’s Rule of 5 characteristics of the top five ranked ligands were retrieved from ZINC15 and documented in the table below. Lipinski’s Rule of 5 states that an orally active drug has no more than one violation of the following criteria:
• No more than 5 hydrogen bond donors
• No more than 10 hydrogen bond acceptors
• A molecular mass less than 500 Daltons
• An octanol-water partition coefficient (log P) that does not exceed 5
Finally, the highest-ranking ligand, Ligand 11, was inputted into PyMOL Visualization software to visualize binding locations on the protein molecule and find any active polar binding site residues. The protein-ligand visualizations are shown in the figures below and the polar residues involved in docking can be found in the data (Tables 4 and 5) (Figures 6-9).
| H-bond donors | H-bond accptors | Molecular mass (Kda) | Log P | |
|---|---|---|---|---|
| Casearinol A | 1 | 8 | 518.647 | 4.615 |
| Ligand 8 | 1 | 8 | 518.647 | 4.615 |
| Ligand 9 | 1 | 8 | 504.62 | 4.225 |
| Ligand 10 | 1 | 8 | 504.62 | 4.225 |
| Ligand 11 | 1 | 8 | 504.62 | 4.225 |
| Ligand 12 | 1 | 8 | 518.647 | 4.615 |
Table 4: Lipinski's Rule of 5 chemical properties of five highest ranked.
| Protein | Ligand | Active polar residues |
|---|---|---|
| E-selectin | Ligand 11 | TYR-44, FUC-4, GLU-80, GAL-2 |
| P-selectin | Ligand 11 | E107, D106, Y94, N105, E92 |
| VCAM-1 | Ligand 11 | VAL-61, PHE-121, GLY-27, GLU-150 |
| ICAM-1 | Ligand 11 | ARG-166, GLN-181, ASN-175, NAG-1, NAG-2 |
Table 5: Active polar site binding residues of Ligand 11 (ZINC000146747094) to each cell adhesion molecule.
Figure 6: PyMOL 3-dimensional visualization of Ligand 11 attachment to E-selectin.

Figure 7: PyMOL 3-dimensional visualization of Ligand 11 attachment to P-selectin.
Figure 8: PyMOL 3-dimensional visualization of Ligand 11 attachment to VCAM-1.

Figure 9: PyMOL 3-dimensional visualization of Ligand 11 attachment to ICAM-1.
After performing the simulation and recording data, it was determined that the hypothesis, that potent inhibitors of cell adhesion molecules involved in lung cancer progression would be found and satisfy Lipinski’s Rule of 5, was supported through evidence from the data. The results revealed that Ligands 8, 9, 10, and 12 had binding affinities greater than controls for E-selectin, P-selectin, and VCAM-1, demonstrating high binding potency. However, the results further showed that Ligand 11 (ZINC000146747094) had binding affinities greater than the two control groups across all four cell adhesion molecule protein groups, with affinities of -8.89 kcal/mol, -7.21 kcal/mol, -7.61 kcal/mol, and -3.67 kcal/mol for E-selectin, P-selectin, VCAM- 1, and ICAM-1, respectively. Moreover, Ligand 11 consistently ranked either first or second among all 13 ligands and controls in terms of binding affinity, indicating it had the strongest binding potential of all chosen ligands. Additionally, it can be seen from (Figure 5) that the five highest ranked ligands had structural similarities to Casearinol A, while from (Table 3) it can be seen that the Manassantin A analogs, which had structural similarities to Manassantin A, had significantly lower binding affinities than the Casearinol A ligands. Moreover, this shows that the differences in binding affinities between ligands is a direct result of the structural differences between the two controls: these chemical variations between molecules may result in increased affinity due to stronger intermolecular forces between the ligand and protein, such as hydrogen bonding. Furthermore, due to these structural differences, the Casearinol A analogs had higher binding affinities than the Manassantin A analogs across all four protein groups.
Nevertheless, this study effectively pinpointed ligands with high inhibition potential of four main cell adhesion molecules involved in lung cancer progression. Moreover, Ligand 11 demonstrated higher binding potential than both controls, indicating that it could potentially be used in a dose dependent manner to inhibit multiple cell adhesion molecules simultaneously, thus decreasing the likelihood of SCLC attachment to endothelial cells and delaying its progression. From a therapeutic standpoint, identifying a potent inhibitor to multiple cell adhesion molecules simultaneously has many advantages. Firstly, having a single, potent inhibitor eliminates the need for multiple treatments and the need for various concentrations of inhibitors. Furthermore, this decreases the likelihood of treatment rejection and makes treatments easier to administer. Additionally, from (Table 4), Ligand 11 has orally active characteristics and satisfies Lipinski’s Rule of 5. In human trials and later stages of testing, these advantages are essential to therapeutic treatment, as the nature of these aggressive, metastatic tumors require treatment in the most efficient way possible.
In addition to identifying potent inhibitors to various cell adhesion molecules, this study also pinpointed specific binding site residues on each of the cell adhesion molecules that can potentially be targeted for future experimentation. After docking, the proteins and Ligand 11, ranking highest among the others, were inputted into the PyMOL visualization software to understand the molecular interactions between the ligand and protein and to view the 3-dimensional protein-ligand attachment. After viewing the protein on the surface view, the polar binding site residues, which the ligand attached to during docking, were recorded in (Table 5). Moreover, by understanding the active binding residue sites utilized by the ligands, a more targeted-based therapy can be used in the future to filter ligands, by only selecting ligands that inhibit the function of the protein by binding to specific residues. Ultimately, specifying the binding residues of each cell adhesion molecule protein opens up the possibility of future targeted-based therapy, which is essential to the development of treatments for SCLC.
Although further experimentation would need to be performed, blocking the communication between endothelial cells and SCLC tumor cells is essential to preventing the extravasation of a secondary tumor site, thus delaying metastasis. By using ligands to block the receptors that mediate transmigration, SCLC cells become unable to attach to endothelial cells and remain in the bloodstream, thus opening the possibility of tumor resection through surgery. To further test this, Ligand 11, along with the other four potent ligands, could be tested in vitro using cell cultures, by first blocking cell adhesion molecule receptors on endothelial cells and then testing SCLC adhesion to endothelial cells through a cell adhesion assay, to see if inhibition had an effect on SCLC attachment. Once the efficacy of these ligands have been confirmed in vitro, the inhibitors could be tested in vivo for toxic side effects and could eventually be used as potent inhibitors to cell adhesion molecules not only in Small Cell Lung Cancer models, but in various metastatic diseases that involve endothelial communication.
In using the Auto Dock 4.0 molecular docking simulation, a few limitations need to be considered. Firstly, the main limitation of this study was the use of a blind-docking procedure, which essentially uses a grid box to allow the ligands to dock onto any residue site on the protein, rather than a specific, known site. The reason for this was due to the relatively little literature on cell adhesion molecule binding pockets utilized by SCLC cells. In the future, when more information is collected regarding specific binding site residues, a target-based docking procedure can be performed, allowing all 50 trials to dock into a specific pocket. Additionally, with the PyMOL Visualization, the biding residues locations of Ligand 11 attachment to each cell adhesion molecule were found; in the future, a target-based docking procedure can be carried out, specifying the binding residues recorded in (Table 5) rather than the entirety of the protein molecule to receive a more accurate binding affinity. Furthermore, this will potentially lead to a much higher binding affinity value, providing more statistical significance to the data. Additionally, another limitation involves the accuracy of the program itself: since molecular docking is only a simulation of protein-ligand interactions, further tests need to be carried out to confirm binding efficacy in vitro. Nevertheless, even with these limitations, ligands with significantly higher binding affinities than known cell adhesion molecule inhibitors were identified, showing support for the hypothesis.
Ultimately, this study effectively identified natural, potent inhibitors to cell adhesion molecules involved in Small Cell Lung Cancer metastasis. The information from this study is useful to future Small Cell Lung Cancer research because it employs a cost-effective method, using software and machine-learning based programs, to find a potential treatment for the main cause of cancer deaths: metastasis. By utilizing docking simulations, this study considered factors such as chemical structures, various binding residue sites, and orientations of molecules to accurately predict the binding affinities of various ligands. Without this study and its use of biotechnology, hundreds of thousands of dollars would be wasted on cell culture testing, with almost no guarantee that a potent inhibitor will be found. Using in silico molecular docking, multiple inhibitors were identified, providing useful information to pharmaceutical companies who can now further test these molecules in vitro and in vivo, which much higher likelihoods of success.
Citation: Akshay P. Kulakarni (2021) A Molecular Docking Approach to the Drug Inhibition of Endothelial Cell Adhesion Molecules Associated with Small Cell Lung Cancer Progression. Drug Des. 10:187.
Received: 24-Jun-2021 Accepted: 08-Jul-2021 Published: 15-Jul-2021 , DOI: 10.35248/2169-0138.21.10.e187
Copyright: © 2021 Kulakarni AP. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.