
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2022)Volume 8, Issue 3
Object: Explore the specific function of CCR7 in immune mechanism of LIHC, and constructed CCR7-related Immune Prognostic Signature (IPS) for LIHC patients.
Methods: The RNA-seq data of LIHC was downloaded from TCGA dataset and the samples were divided into CCR7_H and CCR7_L group. Then, ssGSEA analysis, immune microenvironment analysis, expression level analysis of HLA genes and check point genes were conducted. The Differential Expression Immune Genes (DEIGs) were conducted and LASSO Cox was applied to construct CCR7-related IPS. A novel nomogram was constructed to predict survival rate of LIHC patients.
Results: The Immune score, Stromal score and ESTAMATE score are higher in CCR_H group, while tumor purity higher in CCR_L group. In CCR7_H group, the HLA genes and immune checkpoint genes have higher expression levels. The CCR7_H groups have a favorable prognosis than CCR7_L group. There are 903 DEIG’s were identified. The DEIGs mainly enriched in complement activation, adaptive immune response, T cell activation, lymphocyte differentiation and cytokine-cytokine receptor interaction. The IPS consists of 10 genes, including GHV4-59, SCML4, AKR1B10, LINC00426, TRGC1, F2RL2, TRBV10-3, SAMD9L, SLC4A10 and ROR2. Univariate and multivariate Cox analysis showed that the IPS was an independent prognostic factor of LIHC.
Conclusion: The CCR7-related IPS and nomogram were constructed and provided for LIHC patients to predict survival rate. This study provided a novel way to analyze the prognostic effect of CCR7 expression from the perspective of immunology.
LIHC; CCR7; Immune Prognostic Signature (IPS); Immune microenvironment; Nomogram
Liver Hepatocellular Carcinoma (LIHC) accounts for 85%-90% of primary liver cancer cases, and leads to poor survival with high morbidity and mortality [1]. Because of the delayed diagnosis and limitation of current therapeutic strategy, there are only 18% LIHC patients can survival 5 years after diagnosis [2]. LIHC is characterized by high degree of malignancy, insidious onset and rapid progression, and most patients are often diagnosed in advanced stage [2]. LIHC can be managed by various treatments, including surgical resection, radiofrequency ablation, chemotherapy and radiotherapy [3]. Targeted therapy and immunotherapy have made breakthroughs in recent years, but still with many unsolved problems and need further study [4].
Liver is the largest immune organ of human body, and there are various immune cells. LIHC is typically inflammatory associated tumor with autogenic immunogenicity, and immune tolerance and flight-avoidance mechanisms play an important role in this cancer progression [5]. The dynamical system was reported constituted by cancer cells, immune cell, non-tumor cells and extracellular matrix in tumor microenvironment, which promotes the progression of LIHC base on that composition of the system interact by producing growth factors, cytokines and chemokines [6]. Chemokines, one member of the cell super-factor family, work as attractants in cells migration, and involve in angiogenesis, metastasis, tumor cell growth and invasion [7]. Chemokines are reported promote tumor directly or perform its’ function of immune cell mediation, as well as prevent apoptosis of tumor cells [8]. In addition, the chemokine ligand-receptor interaction perform functions involve in surveillance and inflammation [9].
Chemokine receptor 7 (CCR7), one of G protein-coupled receptors, contributes to tumor neovascularization and accelerates tumor cells proliferation and migration in LIHC [10]. Study report that CCR7 play important roles in lymphatic metastasis, which is closely related to poor prognosis of LIHC patients [11]. CCR7 also was reported participate in epithelial-mesenchymal transition of LIHC cells via impeding histone deacetylase [10]. Besides, regulatory T-cells are key cells that play an immunosuppressive role in the immune microenvironment of LIHC, and it’s function is closely related to CCR7 and SLC (secondary lymphoid-tissue chemokine) [12]. CCR7 can also up-regulate Bcl-2 expression to protect CD8+ T-cell form apoptosis and promote CD3+ T-cells to inhibit proliferation of tumor cell [13]. Therefore, CCR7 was supported to influence immune microenvironment and correlate to prognosis of LIHC patients.
In this study, the LIHC expression data were downloaded from TCGA (The Cancer Genome Atlas) database, and the samples were divided into CCR7 high-expression group (CCR7_high) and CCR7 low-expression group (CCR7_L). The differences between those two groups were evaluated by calculating the level of ssGSEA (single-sample gene-set enrichment analysis) to 29 immune markers. The Differentially Expressed Immune Genes (DEIGs) were also identified between CCR7_H and CCR7_L groups. Then, the prognosis model was constructed based on LASSO regression for LIHC patients, as well as the nomogram model.
Gene expression datasets and data processing
There are 374 RNA-seq data and 377 clinical data of LIHC samples were downloaded from TCGA website (https://portal.gdc.cancer.gov/repository). RNA-seq data was normalized for FPKM values by DEseq2 package of R software. We divided those samples into CCR7_H and CCR7_L groups based on the expression of CCR7.
Immunogenomic analysis and survival analysis
The enrichment levels of the 29-immune signature in each sample were quantized using the ssGSEA score [14,15]. Then, the immune cell infiltration level (immune score), stromal content (stromal score), ESTIMATE score and tumor purity were calculated for each LICH samples [16]. The comparison of expression of HLA genes and immune checkpoint genes between CCR7_H and CCR7_L were conducted by ANOVA test for further analysis. In addition, the overall survival of CCR7_H and CCR7_L in LIHC patients was compared. The log-rank test was used in order to calculate the significance of survival time differences with a threshold of P-value<0.05.
Differential expression analysis and functional enrichment analysis
The differential expression analysis between CCR7_H and CCR7_L genes were analyzed using limma package of R software with the threshold of |log fold change (FC)|>1 and adjusted P values<0.05 [17]. Then, DEIGs were pooled into online tools Metascape (https://metascape.org/gp/index.html#/main/step1) and DAVID (https://david.ncifcrf.gov/) for gene annotation, visualization and providing gene attributes ADDIN EN.CITE [18-21].
Construction and test of the Immune Prognostic Signature (IPS)
The LIHC samples were divided into train set and test set stochastically. The DEIGs in the train set were pooled into LASSO Cox regression to establish immune prognosis signature [19,20]. To estimate a risk score for each patient, an IPS was established by weighting the Cox regression coefficients. Using survminer package of R software, LIHC patients were divided into low-risk and high-risk group. Then, the survival curve and risk curve, especially ROC curve were calculated to evaluate the sensitivity and specificity of this model [21]. The Area Under the Curve (AUC) values from the ROC curves was calculated and testified the prognostic prediction ability of this IPS in the test set synchronously. The concordance index (C-index) was used to evaluate model discrimination.
Development of the nomogram
The independent prognostic ability of the IPS was assessed by univariate and multivariate Cox analyses. Based on the application of the result of Cox analyses, we used the rms package of R software to develop a novel nomogram. On the purpose of determining correction range, calibration plots of observed vs. predicted probabilities of 1-3 and 5-year Survival were also figured.
Immunogenomic analyses between CCR7_H and CCR7_L
The ssGSEA score was conducted to analyze the 29 sets of immune-associated genes of LIHC samples, which represent levels of immune cells, functions or pathways. The ssGSEA results show that in CCR7_H group immune cells, functions and pathways enriched. The Immune score, Stromal score and ESTAMATE score are higher in CCR_H group, while tumor purity higher in CCR_L group (P<0.001). In general, the level of immune status contraries to tumor purity.
The expression of HLA genes and immune checkpoint genes were also analyzed for understand immunogenomic of LIHC. The HLA genes expressed higher in CCR7_H group, as well as immune chick point genes (P<0.001). The survival curve between CCR7_H and CCR7_L was also printed, and the results showed favorable survival rate in CCR7_H group, which has higher immune score and lower tumor purity than CCR7_L group. This means immune compromising condition of LIHC patients in CCR7_H group and potent positive effect of immune checkpoint inhibitors in LIHC patients in CCR7_H group (Figure 1).
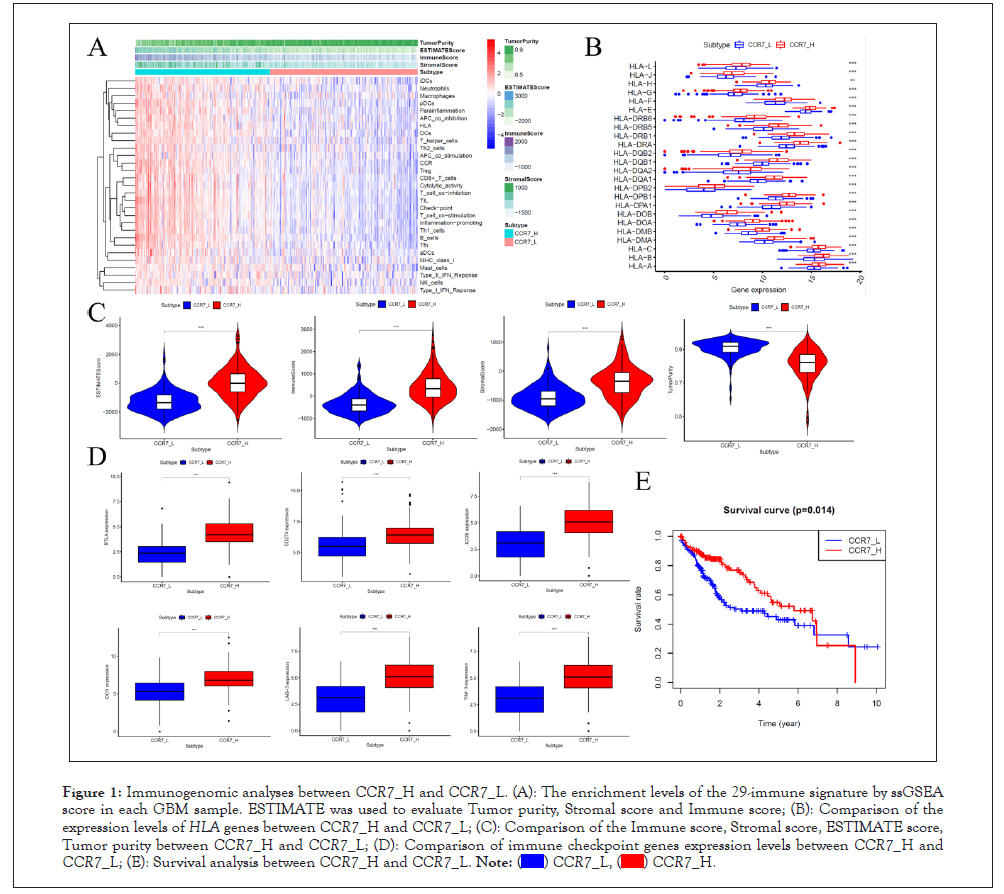
Figure 1: Immunogenomic analyses between CCR7_H and CCR7_L. (A): The enrichment levels of the 29-immune signature by ssGSEA score in each GBM sample. ESTIMATE was used to evaluate Tumor purity, Stromal score and Immune score; (B): Comparison of the expression levels of HLA genes between CCR7_H and CCR7_L; (C): Comparison of the Immune score, Stromal score, ESTIMATE score, Tumor purity between CCR7_H and CCR7_L; (D): Comparison of immune checkpoint genes expression levels between CCR7_H and CCR7_L; (E): Survival analysis between CCR7_H and CCR7_L.  .
.
Identification of differential expressed immune genes
After analyzed the expression levels of immune genes, there are 903 DEIG’s were identified by limma package. Then, the functions and pathways enrichment of DEIG’s was achieved through pooling those genes into Metascape and DAVID website. Results showed that the DEIG’s mainly enriched in complement activation, adaptive immune response, T-cell activation, lymphocyte differentiation and cytokine-cytokine receptor interaction. We also made a network of those terms to clarify the relationship among them, the terms with a similarity of >0.3 was chosen and connected by edges. Besides, when the criteria of no more than 15 items per cluster and no more than 250 items in total were met, the item with the best P value was selected from each of the 20 clusters (Figure 2).
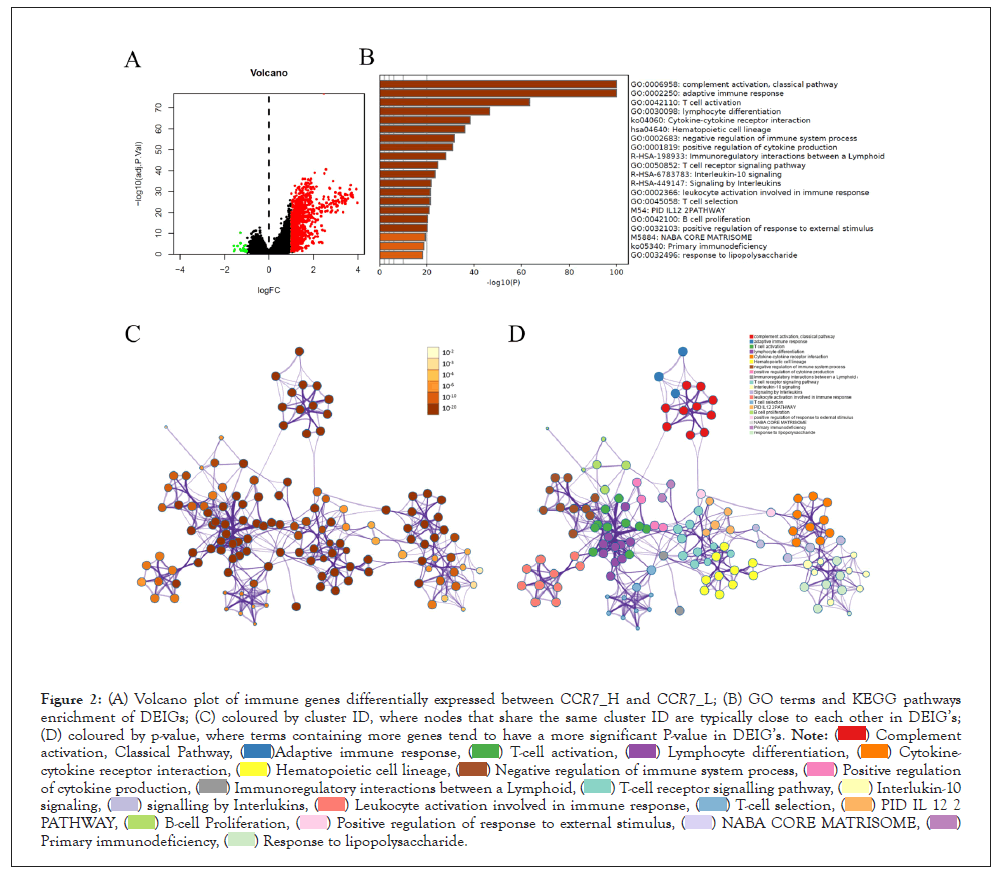
Figure 2: Volcano plot of immune genes differentially expressed between CCR7_H and CCR7_L; (B) GO terms and KEGG pathways
enrichment of DEIG’s; (C) coloured by cluster ID, where nodes that share the same cluster ID are typically close to each other in DEIG’s;
(D) coloured by p-value, where terms containing more genes tend to have a more significant P-value in DEIG’s. 
 .
.
Construction of the CCR7-related immune prognostic signature
The samples in train set were performed by LASSO Cox regression and the risk score for patients can be calculated through coefficients in Table 1. The prognostic signature has good model recognition ability with the C-index (0.755). According to the risk score, patients were divided into high risk group and low risk group both in train set and test set. The survival rate of LIHC patients with low risk score have favorable survival rate in train set, as well as in test set. The ROC curve analysis of IPS showed hopeful prognostic ability for LIHC patients with AUC of 0.763 in train set and 0.706 in test set. In addition, the risk curve and expression pattern of DEIGs in IPS were conducted, the result showed patients with high risk score suffer from shorter survival time and higher mortality rate (Figure 3).
| Genes | Co-efficient | HR | HR.95 L | HR.95 H | p-value |
|---|---|---|---|---|---|
| IGHV4-59 | -0.001234 | 0.998767 | 0.997577 | 0.999959 | 0.042554 |
| SCML4 | -0.003202 | 0.996803 | 0.993533 | 1.000085 | 0.056235 |
| AKR1B10 | 5.59E-06 | 1.000006 | 1.000001 | 1.00001 | 0.018331 |
| LINC00426 | -0.015754 | 0.98437 | 0.964057 | 1.005111 | 0.138659 |
| TRGC1 | -0.004025 | 0.995983 | 0.989235 | 1.002777 | 0.245863 |
| F2RL2 | 0.001597 | 1.001598 | 1.000604 | 1.002594 | 0.001623 |
| TRBV10-3 | -0.066678 | 0.935496 | 0.869104 | 1.00696 | 0.07585 |
| SAMD9L | 0.001694 | 1.001696 | 1.000896 | 1.002497 | 3.24E-05 |
| SLC4A10 | -0.026015 | 0.97432 | 0.940706 | 1.009136 | 0.146428 |
| ROR2 | 0.00103 | 1.001031 | 1.000352 | 1.001709 | 0.002893 |
Table 1: The coefficients of genes in IPS.
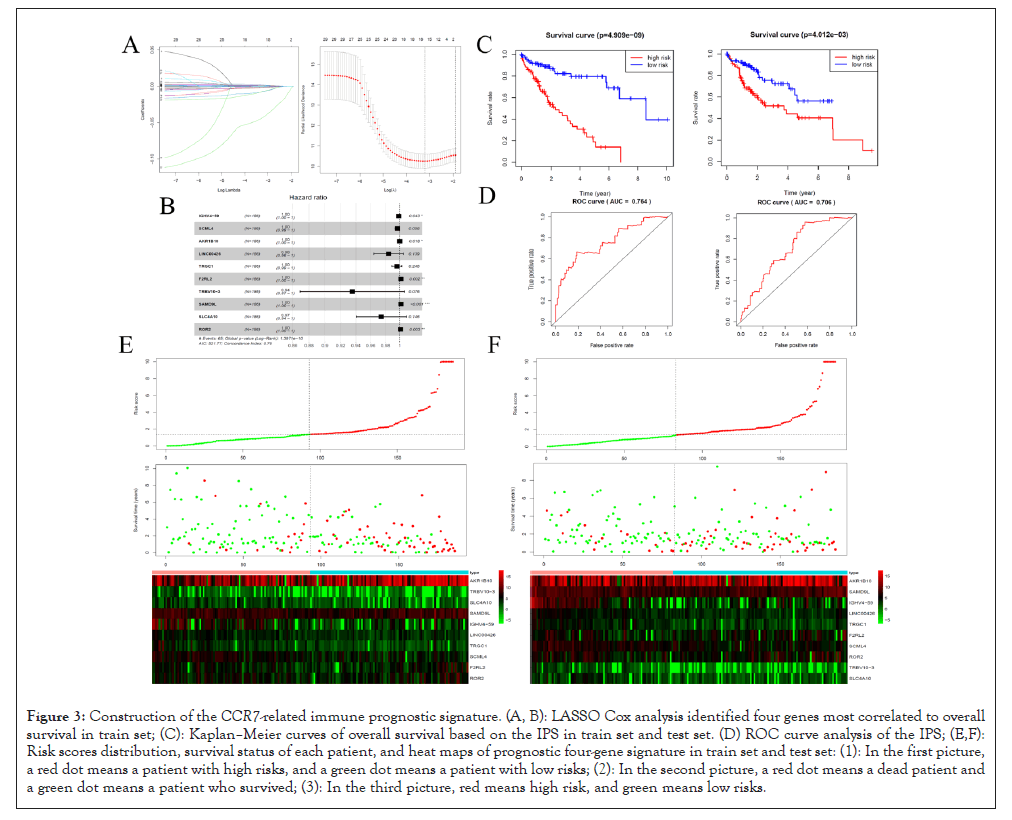
Figure 3: Construction of the CCR7-related immune prognostic signature. (A, B): LASSO Cox analysis identified four genes most correlated to overall survival in train set; (C): Kaplan–Meier curves of overall survival based on the IPS in train set and test set. (D) ROC curve analysis of the IPS; (E,F): Risk scores distribution, survival status of each patient, and heat maps of prognostic four-gene signature in train set and test set: (1): In the first picture, a red dot means a patient with high risks, and a green dot means a patient with low risks; (2): In the second picture, a red dot means a dead patient and a green dot means a patient who survived; (3): In the third picture, red means high risk, and green means low risks.
Establishment an IPS-based nomogram model
Before construct the nomogram model, Univariate Cox analysis and Multivariate Cox analysis were performed. The result showed that the IPS correlated with OS of LIHC patients (Hazard ratio: 1.059 (1.040-1.079), P<0.001) and is an independent prognostic factor (Hazard ratio: 1.056 (1.034-1.078), P<0.001) (Table 2). The nomogram model was constructed based on Multivariate Cox analysis. A good consistency was showed by the 1, 3 and 5 year calibration diagram (Figure 4).
| Cox | ID | HR | HR.95 L | HR.95 H | p-value |
|---|---|---|---|---|---|
| Uni Cox | Age | 1.010933 | 0.99593 | 1.026163 | 0.154063 |
| Gender | 0.81371 | 0.551799 | 1.199937 | 0.298229 | |
| Grade | 1.113202 | 0.861773 | 1.437988 | 0.411626 | |
| Stage | 1.66902 | 1.356537 | 2.053485 | 1.28E-06 | |
| Risk score | 1.059423 | 1.040005 | 1.079204 | 9.61E-10 | |
| Multi Cox | Age | 1.014899 | 0.999657 | 1.030373 | 0.055429 |
| Gender | 0.933891 | 0.623707 | 1.398336 | 0.73983 | |
| Grade | 1.135503 | 0.860345 | 1.498663 | 0.369434 | |
| Stage | 1.591535 | 1.283261 | 1.973864 | 2.33E-05 | |
| Risk score | 1.055509 | 1.03394 | 1.077528 | 2.92E-07 |
Table 2: Univariate and multivariate Cox analyses indicated that IPS was an independent prognostic factor.
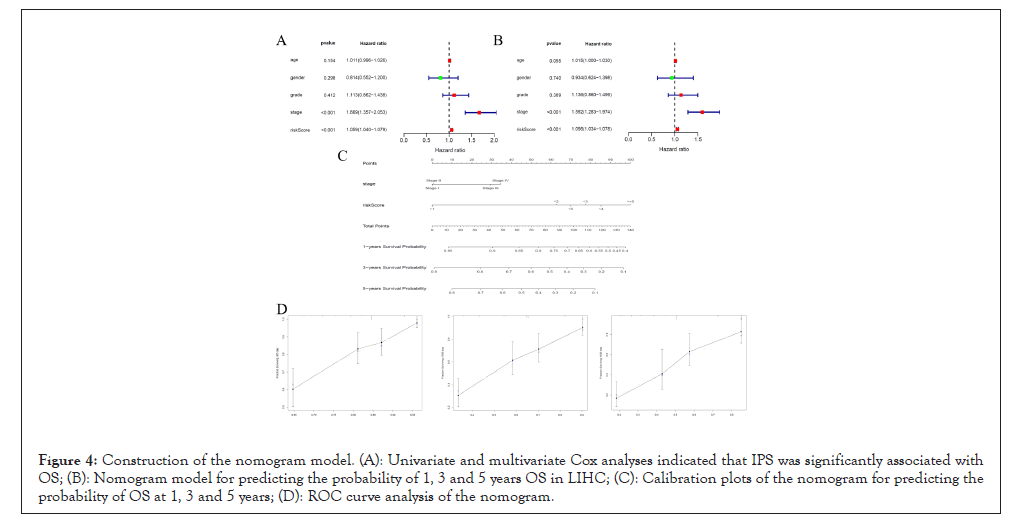
Figure 4: Construction of the nomogram model. (A): Univariate and multivariate Cox analyses indicated that IPS was significantly associated with OS; (B): Nomogram model for predicting the probability of 1, 3 and 5 years OS in LIHC; (C): Calibration plots of the nomogram for predicting the probability of OS at 1, 3 and 5 years; (D): ROC curve analysis of the nomogram.
LIHC is characterized by high degree of malignancy, insidious onset and rapid progression, and contributes to poor survival with high morbidity and mortality [2]. The immune tolerance and flight-avoidance mechanisms were reported play important roles in the progression of LIHC [5], especially the functions of chemokine ligand-receptor interaction [22]. CCR7 was reported contributes to tumor neovascularization and accelerates tumor cells proliferation and migration in LIHC [10]. Therefore, CCR7 was supported to influence immune microenvironment and correlate to prognosis of LIHC patients.
In this study, the ssGSEA results pointed that the CCR7_H group enriched with more immune cells, functions and pathways. The higher immune score, stromal score and ESTIMATE score also were identified in CCR7_H group, as well as lower tumor purity. The expression levels of HLA genes and immune checkpoints genes also perform higher in CCR7_H group. Besides, the survival analysis showed that the CCR7_H group patients have favorable survival rate than CCR7_L group. This means higher immune activity related to CCR7_H, and it can result to a better prognosis. The differential expression analysis was conducted to identify 903 DEIG’s, and the functions and pathways of those DEIGs significantly enriched in complement activation, adaptive immune response, T-cell activation, lymphocyte differentiation and cytokine-cytokine receptor interaction. That explored the molecular mechanism of CCR7 in progression of LIHC.
The IPS of LIHC based on CCR7 was constructed by applying LASSO Cox analysis, and the IPS was evaluated as a good prognostic model with C-index, survival analysis, ROC curve and risk curve analysis. In addition, the Univariate and Multivariate Cox analysis were conducted and fund that the IPS was an independent prognostic factor for LIHC patients. There are 10 genes were identified as hub genes of the IPS, including IGHV4-59, SCML4, AKR1B10, LINC00426, TRGC1, F2RL2, TRBV10-3, SAMD9L, SLC4A10 and ROR2. IGHV4-59 was reported was the most mutation frequent IGHV gene and over expression in chronic lymphocytic leukemia and contributes to progression of this cancer [23] and SCML4 also participate in the mechanism of acute and chronic lymphocytic leukemia [24]. AKR1B10 can protect hepatocytes from damage induced by oxidoreduction and detoxification, the deficiency of AKR1B10 play important roles in development of LIHC especially inflammation related hepatocarcinogenesis [25]. Zhu et al. reported that LINC00426 contributes to immune related signature construction to predict LIHC patients prognosis [26]. TRGC1, T-cell receptor gamma 1 region, was pointed paly roles in functions of T-cells [27]. F2RL2 was reported associated with development of preneoplastic foci in liver and promote initiation of liver cancer [28]. TRBV10-3 was one of subtypes of TRBV10, which related to CD4+ T-cells of primary biliary cholangitis patients [29]. The inactivation of SAMD9L was pointed contributes to progression of B virus-associated hepatocellular carcinoma via facilitation G1-S [30]. SLC4A10 is one of the SLC4 members, which had been proved related to cancer, hypertension and drug addiction [31]. ROR2, the receptor tyrosine kinase-like orphan receptor 2, can promote secretion of Wnt5a to active Wnt signaling pathway in colon cancer cells [32], which associated with proliferation, migration and invasion of LIHC closely [33].
This study elaborates molecular mechanism and constructs IPS of LICH based on CCR7 expression, provide new ways of thinking for LIHC immune microenvironment studies. However, this study is a retrospective research and still has limitations such as lack of foresight. Besides, the specific functions and action mechanisms of those hub genes of the IPS still need further study.
Based on the differences of immunogenomic profiling between CCR7_H group and CCR7_L group, this study constructed the CCR7-related IPS for LIHC patients. The expression level of CCR7 related to prognosis of LIHC patients. The IPS is an independent prognostic factor for LIHC. Besides, the nomogram was constructed to predict survival rate of LIHC patients.
This study was supported by the foundation of the Henan Medical Science And Technology Public Relations Project (LHGJ202105534).
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed]. https://doi.org/10.1002/hep.27246.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed]. 2018;371:231-237.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Wang X, Zhou B, Qin L, Kuai J, Yang F, Yang L, et al. (2022) A Novel CCR7-Related Immune Prognostic Signature for Liver Hepatocellular Carcinoma (LIHC) Based on Immunogenomic Profiling. Immunotherapy (Los Angel). 8:191.
Received: 02-May-2022, Manuscript No. IMT-22-17084; Editor assigned: 05-May-2022, Pre QC No. IMT-22-17084 (PQ); Reviewed: 20-May-2022, QC No. IMT-22-17084; Revised: 26-May-2022, Manuscript No. IMT-22-17084 (R); Published: 06-Jun-2022 , DOI: 10.35248/2471-9552.22.8.191
Copyright: © 2022 Wang X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.