Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research Article - (2022)Volume 11, Issue 3
Coronaviruses are single-stranded positive-sense enveloped RNA viruses that caused the ongoing outbreak of coronavirus disease 2019 (COVID-19). This outbreak is highly aggressive and has become a global threat with a rapid spreading rate. Even though coronaviruses were recognized as veterinary pathogens for a long time, due to their wide distribution across different animals, they also found a causative agent of disease for humans when coronavirus strain OC43 was discovered as the causative agent of the common cold in humans. Since that time various strains of coronavirus are discovered to cause disease for humans. The high mutation and recombination efficiency and diversified natural reservoir of coronavirus give the virus a great opportunity to adapt and evolve rapidly. As a result, the virus emerged as a cause of several outbreaks, including severe acute respiratory syndrome coronavirus (SARS), Middle East Respiratory Syndrome (MERS), and COVID-19. As investigations revealed that all the viruses causing these outbreaks have zoonotic origins. However, we know little about the SARS-CoV-2, an emerging virus, which is the causative agent of COVID-19. In contrary to other types of outbreaks, COVID-19 is a pandemic that kills millions of lives in the whole world. Considering its impact various research institutions are working to develop its vaccines using different strategies. Among the strategies, nucleic acid-based is the most promising approach. Based on this approach several DNA and RNA-based COVID-19 vaccines, such as Moderna, Pfizer, and Johnson-Johnson vaccines have been developed and get approval for use. On the other hand, other vaccines such as CureVac and CVnCoV vaccines are waiting for authorization which revealed nucleic acid base COVID-19 development strategy is a better approach.
Coronavirus; COVID-19; Vaccine; Pfizer; Moderna
Coronaviruses (CoVs) were first described in the 1960s as causative agents of the common cold in humans. Since then, different CoVs have been discovered. In 1967, a new CoV strain, OC43 was also discovered. In the last decade’s other CoVs, such as severe acute respiratory syndrome-CoV (SARS-CoV) in 2003, Middle East Respiratory Syndrome-CoV (MERS-CoV) in 2012, and currently in 2019, SARS-CoV-2 emerge with a spectrum of disease severity. The emergence of SARS-CoV-2, provisionally named novel Coronavirus Disease 2019 (COVID-19), in China at the end of 2019 has caused a large global outbreak. This ongoing viral disease outbreak is highly aggressive and has become a global threat with a rapid spreading rate [1].
In this review, we summarize the potential natural reservoir of coronaviruses, the role of genetic mutation, and recombination in coronavirus evolution, coronavirus outbreaks, and COVID-19 vaccine development processes, which may help to develop strategies to block the viral cross-species transmissions [2,3].
Moreover, comparison among SARS-CoV-2, SARS, and MERS based on clinical symptoms, incubation time, mode of transmission, Complete Genome size, natural reservoirs of coronaviruses, mutation rate, and fatality rates are reviewed to provide a better understanding of the relatedness of COVID-19, SARS, and MERS, and the nature and origin of COVID-19.
Coronaviruses
Coronaviruses are single-stranded positive-sense enveloped RNA viruses with 27-32 kb long genome which is the largest genome among all RNA viruses. CoV genome contains 7-14 Open Reading Frames (ORF) encoding polymerase complex, Spike glycoprotein (S), an Envelope protein (E), Membrane glycoprotein (M), and Nucleocapsid protein (N). The structure of coronavirus as described in Figure 1.
Figure 1: Structural details of coronavirus.
Coronaviruses are classified under Coronaviridae family and Coronovirinae subfamily within Nidovirales order. According to a proposal submitted to the International Committee on Viruses Taxonomy, CoVs are classified into Alpha-coronavirus, Betacoronavirus, Gamma-coronavirus, and Delta-coronavirus genera based on genome-scale phylogenies. Among CoVs genera affecting human health belong to the family of Coronaviridae, subfamily Coronovirinae, only Alpha-coronavirus and Betacoronavirus are of interest for human and clinical virologists, because up to now all human diseases caused by coronavirus are caused by either Alpha-coronavirus or Beta-coronavirus [4].
Potential natural reservoirs of coronaviruses
Even though CoVs are host-specific, they can infect humans and a variety of different animals. They were recognized as veterinary pathogens for a long time, due to their wide distribution across different animals, such as dogs, palm civets, camel, pets, poultry, livestock, humans and bats. Thirty years after CoV was recognized as a disease-causing agent in animals, the first Human CoV (HCoV) was discovered in 1965 by Tyrrell and Bynoe. Since that time several other CoVs, such as SARS-CoV in 2002, Human CoV-NL63 (HCoV-NL63) in 2004, HCoVHKU1 in 2005, and MERS-CoV in 2012, have been discovered. Almost all the HCoVs have zoonotic origins. CoVs are found in a diverse array of bird and bat species as natural hosts. As the only flying mammals, bats are known as a natural reservoir of various human pathogenic viruses including Rabies virus, Nipah virus, Hendra virus, Ebola virus, Marburg virus, and influenza virus. Bats are also considered as the reservoir of several emerging viruses, including coronaviruses that can cause SARS, MERS, Porcine Epidemic Diarrhea (PED), and Severe Acute Diarrhea Syndrome (SADS). Bats are known to harbor high levels of CoV diversity. Several strains of CoV detected in bats are genetically similar to MERS-CoV, SARS-CoV, HCoV-229E, and HCoV- NL63 which support the idea that bats have played a major role in CoV evolution. COVID-19, the cause of the current global pandemic, is found to be closely related to SARSlike CoV sequences that were isolated from bats. Based on the phylogenetic analysis of COVID-19 genome, it belonged to the genus beta-coronavirus and displayed the closest linkage with SARS-like coronaviruses from bat [5]. A study conducted in China also revealed that Pangolin CoV is 91.02% identical to SARS-CoV-2 and 90.55% identical to Bat CoV RaTG13 at the genome level which supports the idea that Pangolin is a probable intermediate host of SARS-CoV-2.
The history of coronavirus diseases outbreaks
In the past decades, several CoVs have caused serious health problems. In 2003, a severe form of pneumonia outbreak which was subsequently named SARS was reported for the first time in Viet Nam, and China World Health Organization (WHO). Until this outbreak, CoVs were not considered to be highly pathogenic to humans. It causes severe respiratory diseases in humans. SARS was reported to have a mortality rate of 3-6%, although another report suggests this rate can be as high as 43-55% in people older than 60 years. In 2012, ten years after SARS, the second highly pathogenic novel coronavirus called MERS emerged in Middle Eastern countries. The third severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), later named COVID-19, is also identified in China, in 2019. COVID-19 showed more than 80% and 50% nucleotide identity to SARS-CoV and MERS-CoV respectively [6].
Severe Acute Syndrome Coronavirus (SARS-CoV)
SARS is the first transmissible pandemic disease caused by SARS-CoV first occurred in China. It resulted in 8000 cases of infection and10% deaths worldwide within 8 months. It has a 10-50% mortality rate depending on the age of infected individuals. In aged populations, more than 60 years old, the mortality rate was higher than 50%. SARS-CoV is likely a recombinant virus that originates from horseshoe bats and palm civets due to its ability to undergo genetic recombination [7]. In the year 2005, independently two teams have reported novel CoVs related to human SARS-CoV were reported in horseshoe bats. These findings strengthen the suggestion that bats are the natural hosts for SARS-CoV. Therefore, it is reasonable to assume that at the beginning of the 2002–2003 outbreaks, there was animal-to-human interspecies transmission and later humanto- human transmission efficiency is adapted.
Middle East Respiratory Syndrome Coronavirus (MERS-CoV)
MERS-CoV is a novel coronavirus that emerged in Saudi Arabia in 2012 and spread into the Middle East, North Africa, Europe, United States, and Southeast Asia. Since its discovery, more than 2000 human cases were reported with a fatality rate of 35% (WHO, 2017). Among 2374 laboratory-confirmed MERS-CoV cases reported to WHO from April 2012 to February 2019, the majority of cases (83.52%) were reported by Saudi Arabia (WHO, 2019).
Different pieces of evidence showed that bats are the original host of MERS-CoV. Like SARS, severe cases of MERS can result from a weakened immune system, older age, diabetes, cancer, renal disease, and chronic lung disease [3,8]. MERS-CoV patients have acquired the infection through infected humans, camels, bats and other domesticated animals. However, dromedary camels appear to be the most likely source of animalto- human transmission as an intermediate host. This is supported by the study conducted on camel workers which reveals approximately 50% of camel workers in the Kingdom of Saudi Arabia were infected.
Novel coronavirus 2019 (COVID-19)
COVID-19 was first noticed in late December 2019. It is a highly transmittable viral infection caused by SARS-CoV-2 that emerged in Wuhan, China first and spread throughout the world. At the moment it becomes a world pandemic by spreading rapidly to all the global countries. SARS-CoV-2 is an RNA virus containing plus sense single-stranded RNA genome and belongs to the Coronaviridae family of beta coronaviruses. It is the causative agent of COVID-19 that had more than 95% homology with the bat CoV and more than 70% resemblance with the SARS-CoV for which relatedness the virus gains its name as SARS-CoV-2 [9]. In terms of novelty, SARS-CoV-2 is different from SARS-CoV and MERS-CoV. SARS-CoV-2 is the seventh coronavirus known to infect humans next to SARS-CoV, MERS-CoV, HKU1, NL63, OC43, and 229E, but it is found more contagious than the others. SARS-CoV-2 can have two origins-
• Natural selection in an animal host before zoonotic transfer.
• Natural selection in humans following zoonotic transfer.
Full-length genome sequences of COVID-19 from five patients revealed as SARS-CoV-2 shares 79.6% sequence identity to SARS-CoV and 96% identical to a bat CoV at the whole-genome level. Molecular analyses showed the genetic association between pangolin CoV and SARS-CoV-2 which is found to have 91.02% nucleotide identity to SARS-CoV-2 which enables us to consider Pangolin as novel intermediate SARS-CoV-2 hosts as described in Table 1.
| Coronavirus | |||
|---|---|---|---|
| MERS | SARS | COVID-19 | |
| Outbreak date | April 2012 (WHO, 2018) | February 2003 | December 2019 |
| Incubation period | 2-14 days | 2 -10 days | 1-14 days |
| Typical symptoms | High fever, myalgia, non-productive cough, nausea, vomiting, and diarrhea | Sudden onset of flu-like prodrome, fever, dry cough, non-respiratory symptoms, like diarrhea, myalgia, headache, and chills/rigors (WHO, 2004) | The patient shows various symptoms, usually fever, cough, breathlessness, fatigue, and malaise among others |
| Origin of HCOVs | Zoonotic | Zoonotic | Zoonotic |
| Mortality rate | 35% (WHO, 2017) | 10%-50% | 5-7% |
| Mode of transmission | Close and prolonged contact | Large droplets and surface contamination | Contact with respiratory droplets rather than through the air |
| Reproduction number | <1 | 1.7-1.9 | 2-2.7 |
| Complete Genome size | 29,809 bp | 29,518 bp | 29903 bp |
| Mutation rate | 1.12×103 substitutions per site per year | 0.80 - 2.38 x 10-3 nucleotide substitution per site per year | Still unclear, but believed to be slightly lower than SARS |
Table 1: Comparison of incubation time, origin, mode of transmission, reproductive number, genome size, mutation rate, and fatality rates of MERS, SARS-CoV, and SARS-CoV-2.
Role of mutation and recombination in coronavirus evolution
The diversity of CoV is due to adaptive evolution driven by high mutation rates and genetic recombination. In this regard, there is growing evidence that some RNA viruses, including CoVs, can generate adaptively useful genotypic variation through recombination and mutation. To understand CoV evolution mutation rate is a critical parameter because-
• RNA viruses have a higher mutation rate than DNA viruses.
• Single-stranded viruses have a higher mutation rate than a double-strand virus.
• Viruses with smaller genome sizes have higher mutation rates as compared with viruses with larger genome size. Based on the genome-wide analysis, the evolutionary rate of CoVs is estimated as the same order of magnitude as in other fastevolving RNA viruses.
In comparison with other single-stranded RNA (ssRNA) viruses, CoVs have estimated mutation rates ranging from moderate to high mutation rates and the average substitution rate for CoVs is 1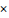 104 substitutions per year per site which is similar to other RNA viruses. Like other CoVs, SARS-CoV-2 used mutations and recombination as crucial strategies in different genomic regions including envelop, membrane, nucleocapsid, and spike glycoproteins to become a novel infectious agent. However, compared to SARS-CoV, the mutation rate of SARS-CoV-2 is lower, which is suggestive that SARS-CoV-2 has a higher level of adaptation to humans [6].
104 substitutions per year per site which is similar to other RNA viruses. Like other CoVs, SARS-CoV-2 used mutations and recombination as crucial strategies in different genomic regions including envelop, membrane, nucleocapsid, and spike glycoproteins to become a novel infectious agent. However, compared to SARS-CoV, the mutation rate of SARS-CoV-2 is lower, which is suggestive that SARS-CoV-2 has a higher level of adaptation to humans [6].
Recombination, a key evolutionary process, accounts for a considerable amount of genetic diversity in natural populations, which is also the most intriguing aspect of CoV replication. Genetic recombination in positive-strand RNA viruses is a significant evolutionary mechanism that drives the creation of viral diversity. It requires the coexistence of at least two genetically distinct CoVs in a single infected cell. CoVs contain a very large RNA genome that undergoes recombination at a very high frequency, nearly 25% of their entire genome. In a study conducted to investigate the origin of SARS, in 2005, seven putative recombination regions located in Replicase 1ab and Spike protein that exist between SARS-CoV and other 6 CoVs: Porcine Epidemic Diarrhea Virus (PEDV), Transmissible Gastroenteritis Virus (TGEV), Bovine Coronavirus (BCoV), Human Coronavirus 229E (HCoV), Murine Hepatitis Virus (MHV), and Avian Infectious Bronchitis Virus (IBV) is found, which is significant evidence about the origin of SARS by recombination events in SARS-CoV genome. A recent publication, discussing the emergence of SARS-CoV-2, suggested that SARS-CoV-2 emerged naturally through the recombination of at least two viruses: A bat Beta-Coronavirus (b-CoV) and a pangolin b-CoV, neither of which normally infects humans. Homologous recombination may occur and contribute to crossâ?species transmission of COVID-19. Because when various CoVs undergo evolution and genetic recombination, mutated CoVs may emerge that may be highly pathogenic to humans. Of course, genetic recombination has been proposed to have a great role in the emergence of new coronavirus lineages, such as SARS-CoV.
Nucleic acid-based vaccines for coronaviruses
The platforms to develop a vaccine against CoVs, include live attenuated vaccines, inactivated whole-virus vaccines, Virus-Like Particle (VLP) vaccines, messenger RNA (mRNA) -based vaccines, DNA-based vaccines, and viral vector-based vaccines. Nucleic Acid (NA)-based vaccines are genetic vaccines that involve direct immunization with RNA or DNA encoding the antigen(s) of interest. In NA-based vaccination, mRNA or plasmid DNA containing antigen encoding gene can be used to trigger humoral and cell-mediated immune responses which can be rapidly and inexpensively produced [8]. NA-based vaccination was reported at the beginning of the 1990s by Wolf and colleagues. Even though the instability of mRNA limited its use, the emergence of plasmid DNA became a promising platform. Currently, several companies are developing NA-based vaccines against Covid-19 and it offers a cost-effective approach to SARSCoV- 2 vaccine development. It is completely a novel approach that is thought to confer greater safety than viral counterparts and can have shorter development cycles. Overall, NA-based vaccine platform development has progressed more rapidly than many other vaccine types, and now makes up some of the most promising COVID-19 vaccine candidates.
RNA vaccines against coronaviruses (COVID-19)
mRNA vaccines were first tested in the early 1990s even though their use was limited because of their instability [9]. Currently using self-amplifying RNA (saRNA) is a newer type of RNA vaccine development strategy. In this method, saRNAs are constructed in vitro to contain the basic elements of mRNA: A cap, 5’ UTR, 3’ UTR, poly (A) tail, larger ORF at the 5’ end that encodes four non-structural proteins, sub genomic promoter and gene(s) encoding the vaccine antigen by replacing the genes that encode the viral structural proteins as shown in Figure 2.
Figure 2: Scheme showing a self-amplifying RNA derived from an alpha virus in which structural genes have been replaced by the gene of interest.
The production of these saRNAs under special conditions (packaging cell lines) leads to the formation of single-round infectious particles carrying RNAs encoding the antigens without the use of a “live” spreading viral infection. STARR™, saRNA with LUNAR®, LNP, and mRNA-1273 are stabilized prefusion SARS-CoV-2 spike protein-encoding saRNA ongoing recruiting vaccines against CoVs. Such COVID-19 vaccines use our normal cell processes to produce safely the SARS-CoV-2 spike glycoprotein antigen, which activates both antibody and cellmediated immune responses.
COVID-19 mRNA vaccines from Pfizer/BioNTech (BNT162b2) and Moderna (mRNA-1273) are saRNA vaccines effective at a low dose (two doses of 30 and 100 mg respectively). Currently, these are the two mRNA vaccines, mRNA-1273 (Moderna) and BNT162b (Pfizer) are the nucleic acid-based vaccines approved for emergency use in many countries that can produce the spike protein from SARS-CoV-2. Both the Pfizer and Moderna vaccines are mRNA-based vaccines that initiate cells to make the SARS-CoV-2 S protein, to generate an immune response. While Moderna is reported to have an overall efficiency of 94.1%, the Pfizer vaccine is reported to have overall effectiveness of 95% in preventing COVID-19. Another COVID-19 Vaccine that gets authorization is the Johnson and Johnson COVID-19 Vaccine, developed by the Janssen Pharmaceutical Companies of Johnson and Johnson. However, this vaccine is a Vector-based COVID-19 vaccine. At the moment, there are many COVID-19 vaccines, like CureVac, CVnCoV, waiting authorization which is also mRNA COVID-19 vaccine candidate that utilizes nucleotides without chemical modifications in the mRNA [10].
DNA vaccines against coronaviruses (COVID-19)
DNA vaccines, also known as nucleic acid vaccines, are plasmid DNA that encodes immunogens or immunogens which are expressed in the eukaryotic organism. In this method, the nucleic acid segment is integrated into a bacterial plasmid carrier that contains the encoding segment for the antigen, plus a promoter and other residual segments from the virus or bacteria of origin. They protect direct injection of plasmids encoding antigens to generate a broad immune response. DNA vaccines deliver coronavirus genes to the human cells and almost all DNA vaccines being tested in clinical trials for COVID-19 use the S protein as the antigen.
DNA based vaccination principle depends on the translocation of DNA into the nucleus of cell where the target gene transcription and translation into the antigen is initiated. Frequently plasmids are used as vectors. Once inside our cells, DNA encoding the SARS-CoV-2 spike glycoprotein is released from the viral vector in the cell’s nucleus where the body’s cellular machinery makes a transcript called mRNA. This transcript is then released into the cytoplasm of our cells where it is used to make the SARS-CoV-2 spike glycoprotein antigens. The DNA fragments, mRNA transcript and viral vector are then rapidly broken down and disposed of by cells. Next, SARSCoV- 2 spike glycoprotein antigen is temporarily displayed on the surface of our cells, where it is recognized as foreign and activates T and B cells of the immune system as shown in Figure 3.
Figure 3: Schematic diagram of COVID-19 DNA vaccine expressing soluble SARS-CoV-2 S protein (SDTM) or full- length SARS-CoV-2 S protein (S).
DNA vaccines are considered superior to mRNA vaccines in terms of formulations needed to maintain stability and delivery efficiency. Nevertheless, DNA vaccines must enter the cell nucleus, where they are subject to the risk of integration and mutation in the host genome. Moreover, the application of viral vector to transfer the gene encoding the target protein, as in the case of Ad5 with the expression of S protein can increase some anxieties associated with immunity against virus in humans [2]. Up to 20 March 2021, there are over 90 vaccine candidates in clinical development, with 22 of them undergoing Phase III clinical trials, 32 in Phase II trials, and 44 in Phase I trials. Up to now, there is only one Vector-based (Johnson and Johnson COVID-19 Vaccine) approved DNA vaccine for COVID-19 even though so many DNA vaccines are under investigation at the clinical level. However, DNA vaccines require complicated delivery systems and generally higher doses and are more difficult to produce [5].
As discussions and findings of different researchers reveal the origin of both SARS-CoV and MERS-CoV were from animals and have been transmitted to humans from the reservoirs. This led to the hypothesis that SARS-CoV-2 may also be originated from animals. Like other coronavirus outbreaks such as SARS and MERS, bats are the most likely natural host of COVID-19, even though identifying the exact bat species that serves as the natural host and the intermediate host of the CoV are remains unclear.
As different researchers’ findings indicate the novel coronavirus (COVID-19) may evolve from the pre-existing coronavirus strains through genetic recombination and mutation and gain a higher reproduction number and transmission capacity as compared to other coronaviruses. In general, tracking the origins of SARSCoV, MERS-CoV and SARS-CoV-2 have important implications to the control and prevention of the pandemic, to develop efficacious drugs, vaccines, and therapeutics specific to COVID-19.
Currently, the foundational knowledge from previous nucleic acid vaccine developments has enabled the astonishingly rapid production of a nucleic acid-based vaccine to combat COVID-19 in 2020. As a result, several nucleic acid-based vaccines have been developed to fight COVID-19.
Some of effective nucleic acid-based vaccines approved for used include Moderna, Pfizer, and Johnson-Johnson vaccines. Based on the progresses achieved to develop COVID-19 vaccine, nucleic acid base COVID-19 development strategy is a better approach. Moreover, the experience and lesson from the current COVID-19 vaccine development can open the way to develop vaccines and therapeutics for other viral disease such as HIV/ AIDS and other infectious diseases; and help to improve the future preparedness to combat such outbreaks.
Conflict of interest
We wish to confirm that there are no conflicts of interest associated with this publication.
Funding
There is no financial support for this work.
Authors' contributions
Mr. Tekeba Sisay prepared the draft of the manuscript and Professor Nega Berhane revised and edited the manuscript.
Acknowledgments
We thank Mr. Mesfin Tsegaw for comments on the manuscript.
Citation: Sisay T, Berhane N (2022) A Research on Coronavirus Natural Reservoirs, Pandemic History, and Nucleic Acid-Based Vaccine Development Progress. 11:234.
Received: 28-Apr-2022, Manuscript No. VMID-22-51691; Editor assigned: 03-May-2022, Pre QC No. VMID-22-51691 (PQ); Reviewed: 17-May-2022, QC No. VMID-22-51691; Revised: 24-May-2022, Manuscript No. VMID-22-51691 (R); Published: 31-May-2022 , DOI: 10.35248/2161-0517.22.11.234
Copyright: © 2022 Sisay T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.