Journal of Antivirals & Antiretrovirals
Open Access
ISSN: 1948-5964
ISSN: 1948-5964
Review Article - (2021)Volume 13, Issue 3
The world is amidst a public health crisis as the pandemic has shook us to the core. The COVID-19 caused by the novel SARS-CoV-2 is of zoonotic origin and this tries to explain what could have been the possible proximal origins for the disease in humans. Our review aims at addressing the question like what structural or genomic vicissitude enabled the viral outbreak across genera and so efficiently infect the human populace across the globe. We also try to discuss the prospect of drug repurposing and scope for vaccine development considering the rapid genome modification of the virus. Another finding lies into the action of pre-existing drugs when they are applied in combination and probably that shades some light on the therapeutic approaches. Several investigations have been performed but we are still in search of a novel antiviral drug. With that vision, our focus shifted on the evaluation of existing drugs with positive response against the novel corona virus. We also try discussing certain trends including increased immunity to the disease in the population from a particular geographical area.
SARS-CoV2; ACE-2 receptor; S-protein; Zoonotic; Drug repurposing
In December 2019 a novel coronavirus, SARS-CoV-2 was identified to be responsible for inducing an acute coronavirus disease associated with fatal pneumonia like symptoms [1]. The terminology SARS- CoV-2 is being popularly used in context to the recent outbreak. Severe Acute Respiratory Syndrome Coronavirus-2, shortened to SARS-CoV-2 is the virus which is responsible for causing Corona Virus Disease (COVID-19) [2].This virus is genetically related to the coronavirus that previously caused the SARS outbreak in the year 2002-2003, however they are distinct viruses [3].Over 3 million cases have already been reported worldwide that has resulted in more than 231,251 deaths across the world as on 30th April, 2020. The outbreak occurred initially at Wuhan, Hubei province in China [4]. This exponentially transmissible disease was declared as a pandemic by the World Health Organization (WHO) on 11th March, 2020. According to WHO, the Case Fatality Rate (CFR) has been estimated at around 3.4% of those infected, however the reported number of cases in much lower than the actual number [5]. Typical symptoms of the COVID-19 include headache, dry coughs, fever, anorexia, anosmia, dyspnoea (breathing difficulties) myalgia and pneumonia induced by the virus in more acute cases [6-7]. The acute symptoms generally appear in the immunocompromised section of the population like the elderly, infants and people with pre-existing chronic illnesses like asthma, hypertension and diabetes [8]. The FDA has granted Emergency Use Authorization (EUA) to hydroxychloroquine sulfate for the treatment of COVID-19 patients. Presently there are no established antiviral therapeutics or vaccine available for the treatment. Hence this creates the grave need to develop novel medications that would act as countermeasures to this viral infection in both prophylactic use as well as treatment of infected patients [7]. Several investigational clinical trials have been launched to evaluate the efficacy of certain repurposed drugs, including anti-viral and anti-malarial drugs. According to studies by a French institution, administration of hydroxychloroquine coupled with azithromycin exhibits a significant reduction in the viral load in the infected individuals [9]. Another study in China reported the use of anitimalarial drug chloroquine in the treatment of COVID-19 patients as it reduces viral replication and also the severity of the disease [10]. However, case-control studies regarding these results have been insufficiently powered and the therapeutic dominance by administering these drugs is unclear. Several concerns have been raised over acute potential cardiac complications that the administration of these drugs can cause especially in infected patients. Clinical investigations have been majorly focusing on the repurposing of several previously approved antiviral drugs. A few examples include the viral protease inhibitor-Ritonavir/Lopinavir which was originally discovered to treat HIV, viral protease inhibitor in Hepatitis C-Danoprevir and the antiviral drug used to treat influenza-flavipiravir [11]. The most notable one among these drugs which is currently undergone 10 clinical trials at more than 50 sites globally is Remdesvir, which was initially discovered to treat Ebola virus associated ailments [12]. This antiviral drug has already exhibited in-vitro and in-vivo activities for both MERS and SARS in animal models [7]. Despite all these advancements, a sufficiently powered randomized case-control trial is yet pending which is necessary to conclude definitely on the efficacy of the antiviral potential of Remdesvir or any other therapeutic advancement for that matter.
Coronaviruses are a sizable family of positive-sense single stranded RNA viruses which are mostly enveloped [13]. Coronaviruses can be broadly categorised into three genera namely the Alphacoronaviruses, Betacoronaviruses and the Gammacoronaviruses, of late there has been a temporarily identified new genera known as the Deltacoronaviruses [14]. Although this virus infects a broad range of vertebrate hosts including humans, it is said to have been extensive in bats [15,16]. The variety of diseases that the coronavirus can cause include a huge plethora of disorders in livestock ranging from upper respiratory tract diseases in chicken to enteritis in cattle and pigs and certain hepatic, renal and neuronal infections as well [17]. In higher vertebrates, majorly humans, coronavirus has been linked with causing clement symptoms associated with the upper respiratory tract infections. However in the last two decades there have been multiple outbreaks of acute and occasionally lethal diseases correlating the respiratory tracts, which were later discovered to be caused by certain novel CoVs proven to be pathogenic to the human populace [18]. The characterisation of the novel CoVs proved that they were phylogenetically dissimilar from the previously identified strains of human CoVs. Further studies also suggested the zoonotic origin of this disease as they were originally found in bats and transmitted to humans through an intermittent host in most of the cases. The novel strains host to another occurred with increased efficacy. Infection caused by these novel strains generally caused moderate symptoms in most individuals, but in few others, the responses appeared to be more acute. In a few cases of paramount infections however, it resulted in the death of the individual due to acute respiratory failure brought about by irreparable damage to the alveoli [19].
The SARS-CoV-2 is the seventh coronavirus which is known to infect the human population. Out of these seven, SARS-CoV, MERS (Middle East Respiratory Syndrome), SARS-CoV-2 are known to cause severe diseases whereas NL63, OC43, HKU1 and 229E are analogous to milder symptoms. Three of these strains have been isolated from animals although a lot of ambiguity prevails regarding the characterisation and mode of infection about each of these [20].
SARS-CoV-2 belongs to the genus Coronavirus in Coronaviridae family [21]. Crown-shaped peplomers with a size of 80-160 nm and a positive polarity of 27-32 kb and are pleiomorphic viruses [22]. The genome of the coronavirus usually encodes the following structural proteins: a) (S) the spike protein; b) (E) the envelope protein; c) (M) the membrane protein and d) (N) the nucleocaspid protein (Figure 1) [23-24]. The viral genome, in addition to a few accessory genes includes six Open Reading Frames (ORFs) which are common to all the coronaviruses. Additionally, certain genes from the SARS-CoV-2 genome exhibited less than 80% nucleotide sequence homology to the SARS-CoV. Evidence that suggested the two viruses belonged to the same species was the 94.4% homology in the amino acid sequences of the seven conserved replicase domains in the ORF1ab that was used for the classification of the species [25].
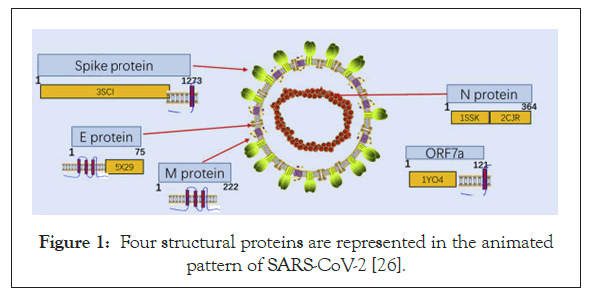
Figure 1: Four structural proteins are represented in the animated pattern of SARS-CoV-2 [26].
After a detailed comparison of the alpha and betacoronavirus, there have been two notable genomic features on the viral spike glycoprotein identified in the novel coronavirus.
The Receptor Binding Domain (RBD) which optimizes the binding of the virus to the human ACE-2 receptor
The most irregular segment in the genome of Coronavirus is the Receptor Binding Domain (RBD) in the spike glycoprotein of the virus. Experimental results from cell culture set ups have proved that six amino acids present in the RBD are essential for binding of the spike glycoprotein to the ACE-2 receptor in human nasopharyngeal pathway (Figure 2) [26,27]. These set of amino acids are also responsible for determining the range of hosts that this novel coronavirus can efficiently infect. With respect to the coordinates based on the SARS-CoV, they are Y442, L472, N479, D480, T487 and Y4911 which correspond to the coordinates L455, F486, Q493, S494, N501 and Y505 on the SARS-CoV-2 genome [28,20]. Five out of these six amino acids vary in the Receptor Binding Domain of SARS-CoV-2 with respect to that of the RaTG13 virus in the bat genome which is supposedly the most closely related virus and also the SARS-CoV [20,29]. With sufficiently powered evidence from biochemical studies it can be concluded that the combination of amino acids in the RBD of SARS-CoV-2 binds to the ACE-2 receptor of ferrets, humans and cats with a considerably high affinity. Although the high affinity binding is something that cannot be ignored, the combination of amino acids in the RBD varies from that which was computed to be optimized for binding to the ACE-2 during analyzing the SARS- CoV binding [30,31]. Hence another notable conclusion from this piece of information could be that the specific sequence in the RBD was a result of the natural selection in the human or human- like host during replication of the virus in initial undetected transmission phase from intermediate host to human or human to human. These observations could conclusively rule out the idea of Figure 1: Four structural proteins are represented in the animated pattern of SARS-CoV-2 [26]. the virus being artificially manipulated in the laboratory [20].
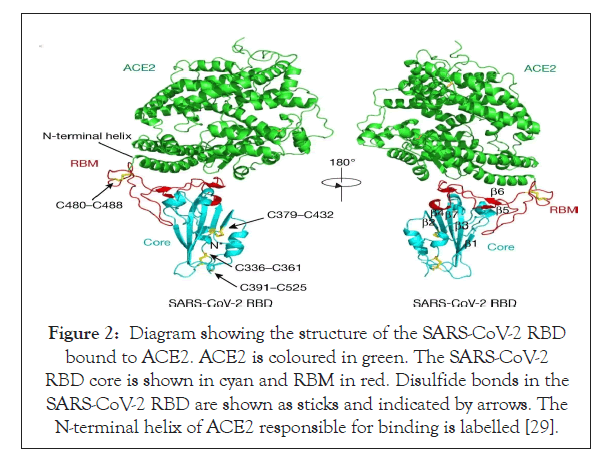
Figure 2: Diagram showing the structure of the SARS-CoV-2 RBD bound to ACE2. ACE2 is coloured in green. The SARS-CoV-2 RBD core is shown in cyan and RBM in red. Disulfide bonds in the SARS-CoV-2 RBD are shown as sticks and indicated by arrows. The N-terminal helix of ACE2 responsible for binding is labelled [29].
Potentially functional polybasic cleavage site for furin protease at the junction of S1 and S2 that occurred via the insertion of 12 nucleotides and also led to the acquiring of three O-linked glycans in the vicinity of that site: The coronavirus membrane contains a transmembrane trimeric spike glycoprotein (S) which plays a significant role in aiding the entry of the virus into the host cell [20,26,32]. The spike glycoprotein harbours two functional domains a) Receptor Binding Domain and b) domain that contains sequences which allow the fusion of the membranes of the virus and the host thus aiding efficient entry of virus into the host cell [33]. The action of the proteolytic cleavage on the S glycoprotein is a major decisive factor whether the virus can jump species and cause efficient infection [34]. Experiments with MERS-like virus showed that the S glycoprotein’s proteolytic cleavage permitted receptor binding of virus from Ugandan bats to humans but hindered entry into the cell. Addition of trypsin in this experiment allowed the S glycoprotein to be adequately cleaved and thus mediated infection of the human host. Such experiment provides evidence for the role of S glycoprotein cleavage as the boundary that allows or hinders a successful zoonotic transfer [35]. After analysing the S glycoprotein, the protein sequence has revealed that it harbours a potential polybasic furin cleavage sequence (RRAR). The RaTG13 isolated from a bat in Yunan shared the maximum nucleotide sequence homology with that of the SARS-CoV-2 genome and even it lacked this furin protease site [36]. Furin protease is an enzyme present abundantly in the nasopharyngeal as well as respiratory tract and hence when the SARS-CoV-2 enters the respiratory pathway, the S glycoprotein is abundantly cleaved after its exit from the epithelial cells. Post cleavage of the S glycoprotein into two subunits S1 and S2 (Figure 3), it gains the ability to infect other cells and mediate entry by fusing the viral cell membrane with that of the host cell [36,37]. Furins also play an important role in determining the pathogenicity of the avian influenza A viruses where the cleavage of a glycoprotein HA is essential for the virus to be able to enter the cell. When the polybasic cleavage site is present at the HA glycoprotein, it converts the otherwise low pathogenic avian influenza virus to lethal forms [38,39]. Addition of the polybasic cleavage site at the junction of the two subunits of the spike protein might be viewed as a certain addition in the functionality of the virus that could have allowed it to jump species. Another notable acquisition of the viral genome could be the gain of O-linked glycans which could possibly give rise to a domain like that of ‘mucin’ that would protect the essential residues present on the RBD of the spike proteins of SARS-CoV-2 [40].
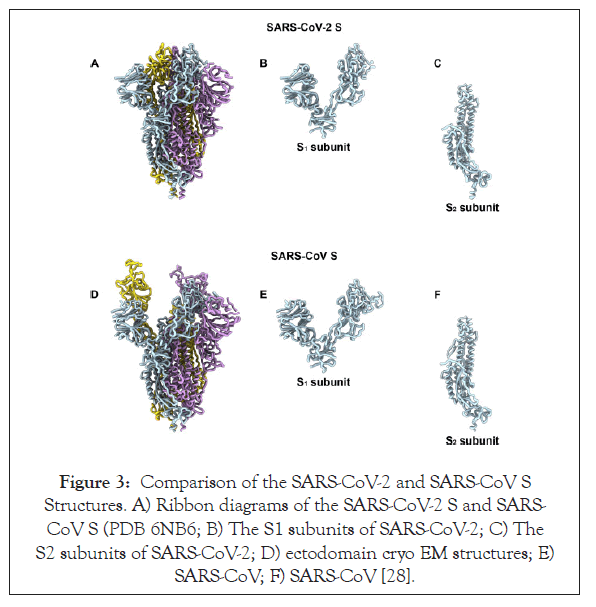
Figure 3: Comparison of the SARS-CoV-2 and SARS-CoV S Structures. A) Ribbon diagrams of the SARS-CoV-2 S and SARS- CoV S (PDB 6NB6; B) The S1 subunits of SARS-CoV-2; C) The S2 subunits of SARS-CoV-2; D) ectodomain cryo EM structures; E) SARS-CoV; F) SARS-CoV [28].
Studies have proved otherwise and demonstrated how natural selection gave rise to this novel variety by means of certain mutations. Understanding of the proximal origin of this pandemic is not only relevant but also urgent in order to understand how this animal virus is capacitated to jump species boundaries and cause acute infections in humans so efficiently. Grave zoonotic events can be prevented or predicted in the future with a broad understanding of the species jump capacitation of viruses [41,42]. Say the SARS-CoV-2 has adequately pre-adapted in a particular animal species already then there are high chances of it manifesting itself in causing another pandemic in the future. On the contrary, if they have adapted themselves sufficiently in the human genome then the chances are that there will be repetitive cases of zoonotic transmissions unless a specific set of mutations occur which had previously occurred. Being able to narrow down the most adjacent viral relative of SARS-CoV-2 will greatly aid clinical studies and more [20].
Two possible hypotheses which can explain the possible proximal origin of the novel coronavirus are
Natural selection occurring in animal host prior to zoonotic transfer: Several cases of COVID-19 were reported to have their origins linked to the Huanan market in China, thus suggesting the presence of the animal host from which the disease spread at that location. Several contradicting views exist regarding the source of the zoonotic transmission. Sampling of the RaTG13 from Rhinolophus affinis (bats) showed a 96.2% homology to the SARS- CoV-2, thus suggesting that reservoir hosts for its progenitor were bats. However, the Receptor Binding Domain in the spike protein of the virus which is essential for binding of the virus to the ACE-2 receptor in humans is different in the bats as the spike diverges in the RBD. This raises concern over unequivocally declaring bats as the reservoir host for the progenitor [41,42].
The coronavirus isolated from Manis javanica (Malayan pangolins), closely resembles SARS-CoV-2. They had been illegally imported in the Guandong province due to the demand of their scales in ancient Chinese medications. Certain pangolins exhibit 100% homology in the Receptor Binding Domain of the spike protein including the six essential residues required for binding to the ACE-2 receptor thus suggesting their role as the reservoir host despite the RaTG13 genome remaining closes to the SARS-CoV-2 genome. There could be a possibility that the virus chose these two organisms or either of these organisms through a process of natural selection and also pre-adapted itself in the genomes of these organisms [43].
The absence of the polybasic cleavage site in the betacoronavirus of bat as well as pangolins raises another plethora of questions. The presence of this site aids the efficient activity of furin protease necessary for the viral infectivity in human hosts. Certain mutations including deletions or insertions can occur at the junction of S1 and S2 of the S glycoprotein thus suggesting that the emergence of the polybasic cleave site by natural selection. For the first theory to be proven right about the virus acquiring the particular sequence in the RBD of the spike protein to bind to the ACE-2 receptor and also acquire polybasic cleavage site, the animal in which the virus pre-adapts need to have a high population density to allow adaptation by natural selection. Additionally, the animal host in which the virus pre-adapts itself also needs to have the ACE-2 encoding gene which would be identical to the human ortholog to a certain extent [20].
Natural selection within the human host succeeding the zoonotic transfer: The other scientifically believable theory about the origination of the virus could be that its progenitor made a species jump and reached the human genome. During a latent phase of undetected human to human transmission, the virus acquired the required characteristics and hence adapted itself to suitably infect the human genome. After the virus has evolved sufficiently to adapt itself it would trigger a cluster of cases with significantly severe symptoms which would lead to a pandemic. All the SARS CoV-2 genome analysis performed so far have confirmed that they all share the above mentioned features thus confirming their arrival from a single source [41]. The fact that pangolins had the identical RBD as that of in the SARS-CoV-2 suggests that it was probably also present in the form of the virus that jumped during zoonotic transmission to humans. This could indicate that the evolution of the polybasic cleavage site occurred only after the virus jumped into humans during adapting itself to natural selection for better furin protease activity to efficiently infect human host. This hypothesis hints at the possible existence of a latent phase prior to the outbreak where undetected human to human transmission occurred [20].
ACE-2 receptor
ACE-2 is the receptor for the SARS-CoV-2 as well as the related human respiratory coronavirus NL63. ACE-2 with homology to the enzyme that has been acted as a major player in the Renin- Angiotensin System (RAS) and hence is a target for hypertension. It is basically a type I metallocarboxypeptidase which primarily expressed in the Leydig cells in the testes, vascular endothelial cells and also renal tubular epithelial cells [44]. Additionally ACE-2 is also expressed in tissues that harbour the SARS-CoV-2 including the gastrointestinal tract, kidney and also lungs. The major substrate for ACE-2 is Angiotensin II as it is degraded by ACE-2 to Angiotensin 1-7 that leads to negative regulation of the RAS. In cardiovascular system and certain other organs ACE-2 is also associated with certain shielding functions [45]. Studies on HeLa cells demonstrated that in humans, civet cats, pigs, mice and Chinese horseshoe bats SARS-CoV-2 uses ACE-2 as the entry point [25]. Yet another interesting finding is in the kidney cell line of monkeys showed that treating Vero-E6 cells (that earlier allowed SARS-CoV replication) with Anti-ACE-2 Antibody inhibited the entry of certain pseudotypes that expressed the SARS-CoV-2 protein [46].
Cleavage by cellular proteases at 2 sites of the S protein is essential for the SARS-CoV entry into the cell by allowing fusion of viral and host membranes and these 2 sites are termed as S protein priming. Out of these two sites, the S protein priming by the serine protease TMPRSS2 is particularly essential for the infectivity of the virus and its ability to spread. Experimental evidence of treating Calu- 3 human lung cell line with camostat mesylate-a serine protease inhibitor hindered the entry of pseudotypes espressing SARS- CoV-2 protein. The physiological role of ACE-2 in the respiratory pathway is currently unknown but it could suggest the role of this enzyme in cleaving certain peptides at the mucosal surfaces in the airway (Figures 4 and 5) [46].
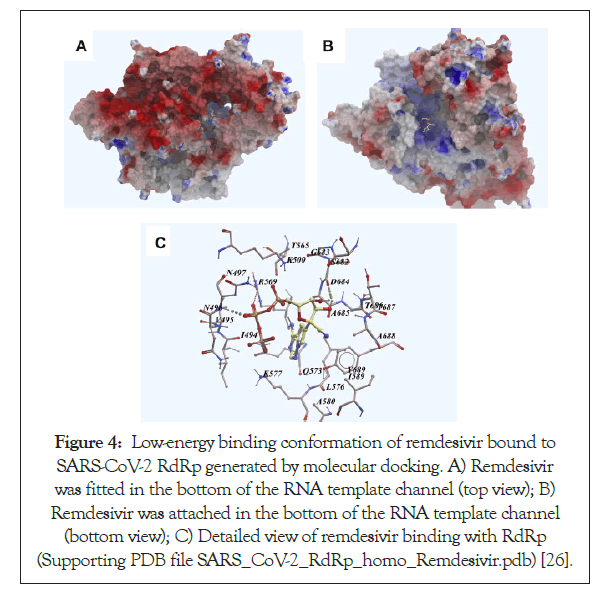
Figure 4: Low-energy binding conformation of remdesivir bound to SARS-CoV-2 RdRp generated by molecular docking. A) Remdesivir was fitted in the bottom of the RNA template channel (top view); B) Remdesivir was attached in the bottom of the RNA template channel (bottom view); C) Detailed view of remdesivir binding with RdRp (Supporting PDB file SARS_CoV-2_RdRp_homo_Remdesivir.pdb) [26].
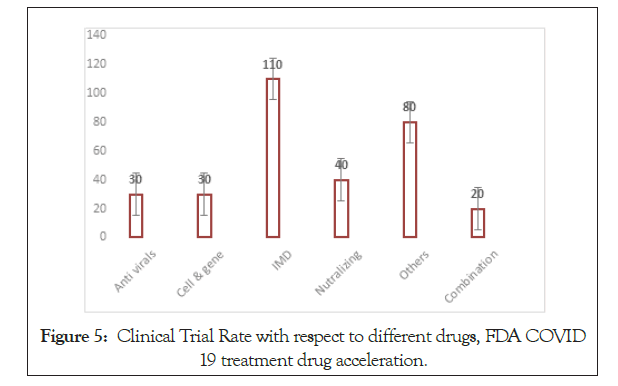
Figure 5: Clinical Trial Rate with respect to different drugs, FDA COVID 19 treatment drug acceleration.
Mode of infection
• The initial step for the viral infectivity is the binding to a receptor in the human host. The Receptor Binding Domain (RBD) in the S1 subunit mediates the binding to the ACE-2 receptor in the human respiratory pathway.
• Next, the S2 subdomain of the S glycoprotein mediates the fusion of the cell membranes of the viral and the human host. The polybasic cleavage site also plays an important role here.
• After this, the S glycoprotein is cleaved into subunits S1 and S2 by the help of furin protease present in the host. After this step the virus gains entry into the host genome and gradually starts replicating by using host machinery [47].
The interaction of the spike proteins with the polarized epithelial cells plays a significant role in the entry of the virus as well as the spread of infection. The cell receptors for the coronavirus are majorly expressed on the apical domain of the polarized epithelial cells as the virus approached the epithelial cells from the apical sides. According to studies performed by immunofluroscence staining, on polarized airway epithelia surface, ACE-2 is more abundantly expressed on the apical side as compared to the basolateral surface. The expression of the ACE-2 receptor is also directly proportional to the stage of differentiation of the epithelial tissue. Experiment demonstrating that ACE-2 expressed in scantily differentiated epithelial tissues was poorly infected while those in sufficiently differentiated epithelial were readily infected, further supported this finding. Among all the epithelial cells, the distribution of the ACE- 2 receptors was found to be maximum on the ciliated epithelium cells which were confirmed by a colocalization experiment with the β-tubulin IV marker of cilia. Hence the infection by the SARS-CoV is positively correlated to the ACE-2 mRNA protein expression and the epithelial differentiation state [48].
COVID-19 is primarily a respiratory illness that affects the upper and lower respiratory tracts of a host through the nasal pathway. There are several ways of collecting samples for testing the presence of coronavirus infection in an individual. Below will be discussed the two most popular techniques used primarily in several south- east Asian countries-RT PCR and the Rapid test kit method. This section shall deal with the grounding principles for each of these detection procedures without commenting on their efficiency.
For the RT-PCR technique, the steps followed are as follows
• Swab is collected from the oropharyngeal region of an individual by either a) injecting saltwater into the nose or b) using a bronchoscope which is a tube that collects sample from the lung. Although it has been concluded that saliva is more sensitive for detection of COVID-19 than nasopharyngeal swabs [49].
• Only certain equipped laboratories can perform this test as the samples need to be maintained in a virus transporting medium and taken to the lab where the tests need to be performed with appropriate safety gear.
• The swab is mixed with certain liquids or buffers such that the virus gets suspended in the liquid.
• Centrifugation is performed in order to separate viral pellets from liquid.
• Viral pellets obtained are then processed in order to isolate the RNA from them.
• Reverse Transcriptase-PCR performed.
• Real Time-PCR performed.
The RNA obtained from the sample is not enough for the detection. Also, the fluorescent dye used to quantify the signals (SYBR green) only binds to double stranded DNA and hence Real-Time PCR has to be performed after Reverse Transcriptase-PCR. The RNA is first treated with the enzyme ‘reverse transcriptase’ that uses the RNA as a template to make complimentary DNA (cDNA). Next, this cDNA is used a template to make a double stranded DNA. A certain signature sequence in the genome of the RNA from the novel coronavirus has been identified as the “signature sequence” which is exclusive to the novel virus. Primers are designed corresponding to these signature sequences and end labelled with SYBR green fluorescent dye. After annealing when the binding takes place and the bases get added, SYBR gets incorporated in the last nucleotides. Hence more will be the amount of signature sequence, more will be the amount of primer binding leading to ds DNA formation and finally leading to even more fluorescence being emitted. If the fluorescence is measured above a certain threshold then the individual from whom the samples were obtained is considered to be infected with coronavirus. PCR is repeated for about 40 cycles by which sufficient amplification would have already occurred to comment on the fluorescence signal emitted and hence the viral infectivity. Only if we see a spike in the fluorescence signal with respect to time, it would suggest a positive result. This whole procedure takes about 8-10 hours and has about 98% accuracy and hence has possibilities of reporting false positives in rare cases. This is not a pathological method to test for infectivity and is more of a laboratory procedure used for research purposes [50].
The other method which is being recently talked about is the rapid test kit method by which an individual could perform a test by himself and obtain results within 10-15 minutes. This process requires following simple instructions and relying on the test kit for the results. First blood sample needs to be obtained after a simple needle prick. The test kit two cavities, one for the input of the blood sample and another for the input of the buffer which is provided in the test kit. The kit has 3 sections-C (control), IgG and IgM. The buffer makes sure that the blood reaches the columns for control, IgG and IgM by a capillary rise technique. When a line is visible at the C section of the kit, it indicates that the test is being run positively and the blood has reached the IgG and IgM sections for calculating the antibodies present in the blood. This test works on the principle of measuring the amount of antibodies expected to be present in the blood of an individual post-infection by the SARS-CoV. This test is not able to provide distinct results for infection by SARS-CoV and SARS-CoV-2. IgM is considered to be the primary antibody to be released immediately after an infection thus suggesting its levels would be high if the individual has been infected recently or is currently under the infection procedure. If the individual testing is in the above mentioned phase, the test would indicate a visible line against the IgM section. However, if the individual had been infected quite a while ago and is already in the recovery phase, there would be lesser amount of IgM in his blood and thus would only see a line against the IgG section. Another notable fact about this test could be that this test considers that each individual testing has been infected by the SARS-CoV at some points in their lives and displays results accordingly.
Several other testing kits, procedures and principles are currently being used around the world and the procedures vary from one country to another.
Therapeutic approach
In a viral disease, no drugs are ultimate. Doctors are trying to reduce the symptoms and conditions through several combinations of drugs. Here is a list of drugs which have proven positive results in experimental set up (Table 1) [51-65].
| Remdesivir | Drugs originally discovered to treat Ebola like viruses |
| It is a single species isomer of the 2-ethylbutyl L-alaninate phosphoramidate which has shown positive results in COVID-19 treatment and has clinical potential. | |
| It is an adenosine analog that inhibits the activity of the RNA-dependent RNA-polymerase and hence blocks the viral replication cycle (Figure 4) [24]. It does so by causing pre-mature termination of transcription when it is incorporated into nascent RNA. After incorporating this drug into the RNA, any more nucleotides cannot be added and hence terminates the transcription [7,51-54]. | |
| Ritonavir | Originally discovered to treat HIV and works by inhibiting the HIV protease enzyme. |
| It is often used in combination with other anti-viral drugs like Lopinavir and sold under the name ‘Kaletra’. | |
| It has shown promising results in marmosets infected with MERS and SARS [55,56]. | |
| Chloroquine and Hydroxychloroquine | Originally discovered as an anti-malarial drug used to treat susceptible infection from several species of Plasmodium. It has also been used for second line treatment of rheumatoid arthritis. It is now being repurposed for antiviral treatment after showing promising results in certain experimental set-ups. However, it shows positive results only when consumed in very large doses which have toxicities of their own. The drug acts by reducing the pH of the endosomes drastically thus affecting viral growth and proliferation. Hydroxychloroquine when administered with azithromycin showed promising results in experiments performed by Gautret et al. However, due to lack of sufficiently powered case-control, the drugs need to undergo more trials [9,57-59]. |
| Darunavir | Originally discovered to treat HIV and was used in combination with Ritonavir and sold as ‘Norvir’. It is a class of protease inhibitors that inhibit the proliferation of the virus within the host [60-62]. |
| It was majorly used to treat patients with a history of retroviral therapies. | |
| Lopinavir | Originally discovered to treat HIV and used in combination with Ritonavir that is being repurposed to treat COVID-19 [63]. |
| Galidesivir | Nucleoside (adenine) analog that was originally discovered to treat the Zaire ebolavirus. It has displayed broad-spectrum anitiviral activity against filoviruses, arenaviruses and coronaviruses. It is being clinically tested to check for potential against SARS-CoV-2 [64]. |
| Favipiravir | Initially used to treat resistant cases of influenza, this is a modified pyrazine analog. This drug functions as a prodrug and works by a novel mechanism to inhibit viral proliferation. This undergoes intracellular ribosylation and phosphorylation to convert to active flavipiravir-RTP. When this is incorporated into the RNA, it prevents viral replication by hindering strand elongation. Due to this ability to function as a broad-spectrum drug resulting from conserved RNA-directed RNA polymerase domain, this is being tested as a potential drug for COVID-19 [65]. |
| Azithromycin | Originally discovered for treatment of respiratory infections, this was approved by the FDA in 1991. It works as a broad-spectrum macrolide antibiotic which has high degree of tissue penetrating potential. When used in combination with hydroxychloroquine, it showed promising results in recovery of patients infected with COVID-19 [9]. |
| Anakinra, tocilizumab, sarilumab, siltuximab | IL 1 and 6 antagonist |
| Adalimumab, infliximab | Anti tumornectrosis factor alpha |
| Gimsilumab, lenzilumab, namilumab | Granulocyte macrophage colony stimulating factor |
| Baricitinib and Ruloxitinib | These are the drug which inhibits the Jak-Stat pathway. Jak-Stat pathway helps in the proliferation of ACE2 receptors. By blocking this pathway, the pathogenesis of SARS-Cov2 can be inhibited. The drugs are in several trials. |
Table 1: List of drugs which have proven positive results in experimental set up.
Considerably higher concentrations of IL-6 have been found to be corresponding with respiratory failures in patients affected by the COVID-19 [66]. Hence, in the clinical approach to design drugs to combat this disease, drugs that ameliorate the inflammatory response produced by the virus are being studied. Additionally another major approach towards drug designing for the ongoing pandemic has been the repurposing of pre-existing anti-viral and anti-retroviral drugs which are already available. One great advantage of repurposing drugs is that it significantly reduces the 10- 17 year time usually required to develop a drug to about 3-12 years. Additionally, under the Emergency Use Authorization (EUA), for Chemical, Biological, Radiological and Nuclear (CBRN) threats such as public health emergencies during such as during SARS- CoV-2 this time period could be reduced to a mere 6 months. Among many other available techniques one of the most practical way to identify anti-viral drugs that could target viral replication could be through high-throughput screening of repurposed drugs [7].
Plasma therapy failure
According to the article of The Hindu of 9th September, 2020 Convalescent Plasma Therapy (CP) is failed in India to benefit in the COVID-19 patient.
Total Hospital included in the trial: 39.
Total patients (n)=464
Time: April-July, 2020
Mortality rate is shown in Figure 6.
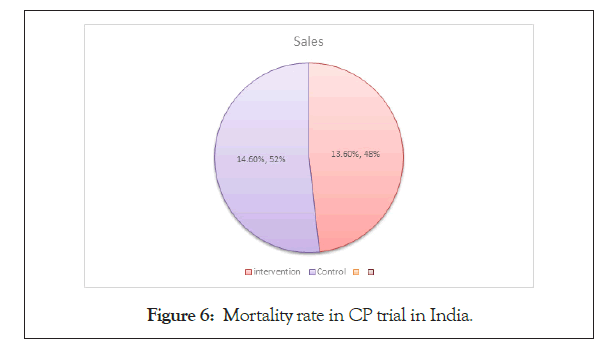
Figure 6: Mortality rate in CP trial in India.
Intervention Arm (n1=235): 34 (13.6%).
Control Arm (n2=229): 31 (14.6%).
Immunopathology and vaccines against COVID-19
Possibly, the only mode of transmission of the disease is from human to human as it spreads by spraying of respiratory droplets like cough or sneeze. The asymptomatic probation period of the virus has been estimated to be around 2-14 days during which it can also infect other individuals. Hence, the average number of humans one person can infect is about 2.2. Study of hospitalized patients have proven elevated levels of release of pro-inflammatory cytokines associated with the SARS-CoV-2 infection like IL-2, IL-7, IL-10, G-CSF, IP-10, MCP-1, MIP-1A, and TNFα [67]. This ‘cytokine storm’ is believed to initiate complications like viral sepsis, lung injury induced due to inflammation and Acute Respiratory Distress Syndrome (ARDS).
Innate immune response: Information regarding the host innate immune response is limited to few studies. One study in Wuhan on SARS-CoV-2 affected patients showed that patients showed a 38% increase in the neutrophil count, 35% reduction in total number of lymphocytes and 52% increase in IL-6. Majority of successful innate immune responses are due to interferon (IFN) type I reaction and its downstream signalling cascade that a) induces adaptive immune response and b) inhibits viral replication. Assuming type I interferon to be of paramount significance in innate immune response, using IFN from mouse model of MERS/SARS can be used as an antagonist to yield protective response.
Adaptive immune response: This could be considered to be the future for vaccine development against SARS-CoV. The “cytokine storm” produced due to the innate immune response induces and orchestrates cell-mediated immune responses [68]. Helper-T lymphocytes mediate the overall adaptive immune response while the cytotoxic-T cells are essential for the killing of the viral cells. Day 9 from the day of onset of infection marks a peak elevation in the levels of IgM while this shifts to peak of IgG by the end of second week. Long lasting specific IgG and neutralizing antibodies can be detected in blood for up to 2 years after onset of infection.
Based on studies during the MERS and SARS outbreak, certain conclusions have been drawn on development of vaccines and their mechanisms. The antigen target selections and the vaccine platform have also been derived from previous studies. As the receptor binding domain is present on the S glycoprotein of the virus, it could be an efficient antigen target for a vaccine. Targeting the S1 region of the S glycoprotein can not only neutralise the effect of the antibodies but also prevent attachment to the ACE-2 receptor and thus inhibit infection. With respect to emerging pathogenicity of pathogens nucleic acid based vaccines, DNA vaccines have proven to be the best platform for delivery [69-70].
This ongoing pandemic has raised several questions and left the entire scientific community baffled. One interesting observation is that the rate of mortality among patients infected with COVID-19 in the SAARC countries is much lesser than that in Western countries such as Italy, Spain, Iran and the USA. For every 1000 people infected in the SAARC countries, about 28 people die while the number goes up to about 76 people in the Western countries. A possible explanation for this geographical anomaly could be the BCG (Bacille Calmette Guerin) vaccine. In several south-east Asian countries this prophylactic vaccine is administered to infants or children to immunize them against tuberculosis. This vaccine could have possibly played some role in providing some sort of immunity against the coronavirus. Among a few other observations is the trend of more number of males getting affected as compared to women. So far there has been no scientific explanation for increased susceptibility of men towards this virus and whether this is a considerable factor is yet to be answered.
A few other important questions could address the potential of the virus to jump species and efficiently infect humans across the globe. A reasonable explanation for this could be certain unique features in its genome including the unique sequence in the receptor binding domain and the polybasic cleavage site. However, which domain of this virus exactly needs to be targeted in order to create a prophylactic or treatment drug is yet unanswered.
Citation: Chatterjee S, Dhara B, Mukherjee D, Mukhopadhyay D, Mitra AK (2021) A Review on the SARS-CoV-2 Mediated Global Pandemic: Proximal Origin, Pathogenicity and Therapeutic Approaches. J Antivir Antiretrovir. 13:220.
Received: 12-Mar-2021 Accepted: 26-Mar-2021 Published: 02-Apr-2021 , DOI: 10.37421/1948-5964.21.13.220
Copyright: © 2021 Chatterjee S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.