
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2021)Volume 11, Issue 2
In general, high grade gliomas have a dismal prognosis. However, recent advances in disease classification, surgical treatment, and adjuvant therapy have increased the overall survival of this patient population. While a significant number of resources is used to improve treatment for high grade gliomas, there are few advancements in technology development towards an accurate non-invasive assessment of response to these therapeutic modalities. The advent of new treatment modalities and especially the increasing number of immunotherapy clinical trials for high grade gliomas necessitate the development of new approaches for accurate and timely assessment of treatment response in brain tumors. Conventional MRI assessment of response to immunotherapy is inadequate to guide treatment leading to a high number of surgical procedures for definitive assessment. In this review, we are outlining the evolution of imaging criteria as well as the advancements in imaging technology and computational analysis to address the challenges of disease monitoring in the setting of immunotherapy for high grade gliomas. We give particular emphasis on radiomics, a new field on image analysis that algorithmically assesses a large number of imaging features for an accurate diagnosis.
Immunotherapy; Advanced imaging; High grade glioma; Pseudoprogression; Radiomics; Clinical trials; Disease monitoring
Glioma is one of the most common and malignant types of primary brain tumors in adults as they account for 28% of all brain tumors and 80% of all malignant brain tumors [1]. Glioblastoma, a high grade glioma, is the most aggressive and refractory brain tumor with a median survival of less than two years [2]. The current standard of care includes surgical resection, radiation, and adjuvant chemotherapy with temozolomide [3]. The majority of patients will experience disease relapse and no therapy currently exists that significantly prolong survival after relapse [4]. In fact, from the time of first between six to nine months [5]. Due to the lack of significant improvement in survival using these conventional therapies, progression or recurrence, median survival duration ranges immunotherapy as a novel therapy is being pursued to improve outcome in glioblastoma patients [6]. Immunotherapy relies on stimulating the host immune system to target and eradicate cancer cells through both innate and adaptive immune responses [7]. By overcoming the immunosuppressive Tumor Microenvironment (TME), immunotherapy has improved clinical outcomes in patients with recurring solid tumors that are resistant to other treatments [8,9]. Different immunotherapeutic modalities include, but are not limited to vaccine immunotherapy, cell-based immunotherapy, and checkpoint inhibitor immunotherapy [6]. For high grade glioma patients, however, these therapeutic options have yet to show significant efficacy [10].
Progress in high grade glioma patient outcomes has been limited not only by the lack of effective treatments, but also by the inherent difficulty in assessing treatment response. Non-invasive, longitudinal imaging has been central to evaluating disease progression and treatment response in neuro-oncology [11]. Historically, structural imaging such as Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) were used to monitor changes in tumor morphology and size in response to conventional treatment [12]. However for a tumor-directed inflammatory response seen in immunotherapy, traditional response criteria and conventional imaging techniques have been poor predictors of therapeutic outcomes [13]. Radiation and chemotherapy for high grade gliomas can lead to an increase in tumor edema, volume, and enhancement shortly after initiating treatment [14,15]. These changes can cause difficulty in detecting disease progression when using conventional imaging techniques and response criteria [15]. This apparent post-treatment increase in tumor size on imaging, followed later by tumor regression and clinical improvement, is known as Pseudoprogression (PsP) [16]. In particular, the inflammatory response and PsP after immunotherapy are often indistinguishable from true Progression of Disease (PD) (Figure 1) [17,18].
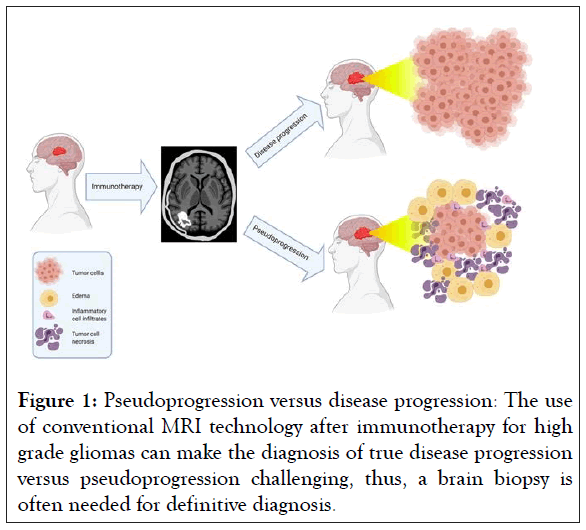
Figure 1: Pseudoprogression versus disease progression: The use of conventional MRI technology after immunotherapy for high grade gliomas can make the diagnosis of true disease progression versus pseudoprogression challenging, thus, a brain biopsy is often needed for definitive diagnosis.
Given the short overall median survival in high grade glioma patients, it is imperative for clinicians to accurately and swiftly distinguish between PsP and PD in order to properly tailor a patient’s treatment plan. Prematurely stopping an effective treatment or maintaining an ineffective therapy will negatively impact outcomes.
This review summarizes the current state and future directions of different non-invasive imaging techniques and criteria used to evaluate treatment response including immunotherapy in high grade glioma patients. Also, novel ways to leverage the imaging data through the field of radiomics is discussed.
Overview of imaging criteria
Various non-invasive imaging criteria have been established over the years in an attempt to create widely applicable metrics for evaluating disease progression and treatment response across studies in neuro-oncology.
Macdonald criteria: Historically, the standard algorithm used to assess oncological disease status and treatment response was the Macdonald criteria [19]. Introduced in 1990, it measured glioblastoma tumor burden by the two-dimensional measurement of contrast-enhancing lesions identified in MRI or CT [19]. It also included metrics to help define Complete Response (CR), Partial Response (PR), Stable Disease (SD), and Progressive Disease (PD) which provided a standard to allow for the comparison of progression-free survival between clinical trials [19]. However, as new therapies were developed, this criteria did not adequately differentiate between PsP and PD [20]. In the MacDonald criteria, a significant increase (at least 25%) in the contrast-enhancing lesion is used as a surrogate marker for PD and often leads to a change in treatment [19]. However, contrast-enhancement is nonspecific and often reflects the passage of contrast material across a disrupted blood brain barrier [21]. Increased enhancement can be caused by a variety of nontumoral processes such as postsurgical changes, seizures, ischemia, radiation necrosis, and treatment-related inflammation [22–25]. Enhancement can also be affected by a variety of factors including changes in radiologic techniques, corticosteroid doses, and the use of antiangiogenic agents [26,27].
The introduction and increased usage of bevacizumab, an antiangiogenic agent, as a treatment for glioblastoma gave rise to the phenomenon known as pseudoresponse [21]. It falsely suggests on imaging that the malignancy responds to treatment and is poorly identified by the MacDonald criteria [13]. The imaging reflects significant reduction of tumor enhancement and edema that is due to a phenomenon known as vascular normalization, in which there is a reduced permeability to contrast agents rather than a true anti-tumor effect [28,29]. Other limitations of the criteria include difficulty in assessing diffuse tumor burden and with low grade tumors that lacked contrast enhancement [30].
RANO criteria
These shortcomings gave rise to the introduction of a new set of standards in 2010: The Response Assessment in Neuro- Oncology (RANO) criteria [21]. To better address the difficulties in differentiating PsP from PD, they included non-contrasting, Fluid-Attenuated Inversion Recovery (FLAIR) MRI and spatialtemporal factors to help differentiate PsP from PD [21]. Furthermore, they did not declare PD if increasing enhancement or new lesions were observed in irradiated target areas of the brain within the 3 months following completion of chemoradiation therapy [21]. The only exception was if there was a new enhancement outside the main radiation field or pathologic confirmation of PD [21].
The criteria also incorporated several metrics such as clinical status, non-enhancing disease evaluation, and usage of corticosteroids to more accurately define CR, PD, SD, and PD [21]. Given the timing at which these criteria were introduced, many of the recent immunotherapy glioblastoma trials utilize RANO criteria [11]. Although several studies have demonstrated the superiority of the RANO criteria compared to the Macdonald criteria, neither fully discriminate PsP and PD [31-33].
iRANO criteria
In 2016, with the rapid expansion of immunotherapeutics under investigation for high grade glioma treatment and the recent development of other immune response criteria for other solid tumors (Immune-Related Response Criteria (irRC), Immune- Related Response Evaluation Criteria In Solid Tumors (irRECIST), and Immune Response Evaluation Criteria In Solid Tumors (iRECIST)), the RANO criteria were updated and adapted to specifically address immunotherapeutic responses [34,35]. They are now called iRANO (Immunotherapy Response in Neurooncology) criteria [35]. Because of the difficulty in differentiating PsP from PD following immunotherapy, one of the main focuses of this criteria was to provide recommendations for management of patients with early progressive changes seen on imaging after beginning immunotherapy [35]. iRANO differs from the RANO criteria by advocating repeat brain MRI after three months if patients demonstrate PD within the first six months of initiating therapy and stable neurologic function [35]. At repeat assessment, if PD is still present, the patient should be classified as having PD from the date of initial noted radiographic progression and treatment modification is recommended [35]. However, if repeat imaging shows stable or improved disease, the immunotherapy treatment should continue [35]. After six months, new or continued worsening is defined as PD while any decreased lesion size indicates an anti-tumor effect [35]. Unfortunately, like the Macdonald and RANO criteria, the iRANO criteria are operator-dependent and rely on two-dimensional measurements (MRI and CT) of enhancing lesions, which are made more difficult with an irregular or ill-defined margins. Furthermore, this watchful waiting jeopardizes patients that have PD to remain on ineffective therapy with its potential toxicity and delaying more effective treatment; conversely, the failure to properly identify PsP can lead to premature discontinuation of an effective immunotherapy.
mRANO criteria
Thus, in 2017, a modified RANO (mRANO) was released [36]. The modifications include the use of contrast-enhanced T1 subtraction maps to increase lesion conspicuity, the use of the post-radiation time point as the baseline for newly diagnosed glioblastoma response assessment, the removal of qualitative non-enhancing tumor assessment requirements, and a treatment-agnostic assessment for identifying PD, PsP, CR, and pseudoresponse [36].
The original study’s primary safety endpoint was freedom from Severe Adverse Events (SAEs) during the procedure and through 30 days after operation [10]. The SAEs were defined based on MVARC criteria [11]. All patients were followed for 1 year with those end points measured at 1, 3, 6, and 12 months. We assessed the New York Heart Association (NYHA) class at baseline, discharge and 1, 3 months.
We assessed MR and mitral valve annular diameter on echocardiography at baseline, intraprocedural period, discharge, and 1, 3, 6, 12 months.
Non-invasive imaging options
Due to inherent limitations in conventional imaging techniques, attention is being directed towards other non-invasive longitudinal imaging options to evaluate high grade glioma patients. The non-invasive imaging techniques can be categorized as structural imaging and functional imaging (Figure 2). Structural imaging includes conventional MRI and CT whereas functional imaging relies on more advanced imaging techniques. Functional imaging provides physiologic and metabolic information that cannot be obtained from structural imaging and includes both physiologic and molecular imaging techniques. Advanced MRI, Positron Emission Tomography (PET), and Single Photon Emission Computed Tomography (SPECT) can be modified and used for both physiologic and molecular imaging. Physiologic imaging evaluates differences in perfusion, fluid diffusion, and environmental conditions to differentiate between healthy and malignant brain tissue [11]. Molecular imaging modalities visualize specific proteins, receptors, or metabolites associated with a targeted pathology, tissue, or cell population [11].
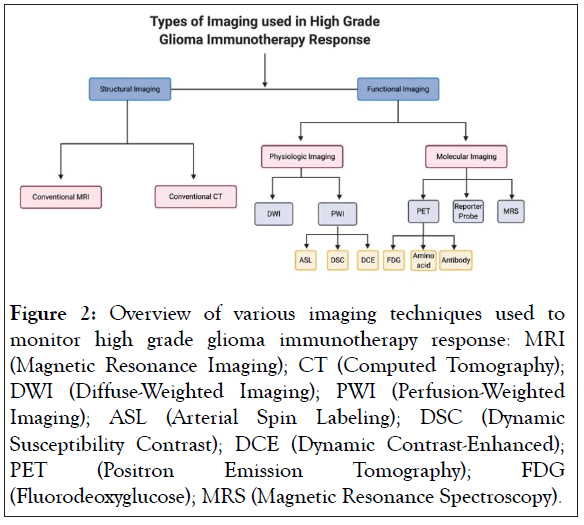
Figure 2: Overview of various imaging techniques used to monitor high grade glioma immunotherapy response: MRI (Magnetic Resonance Imaging); CT (Computed Tomography); DWI (Diffuse-Weighted Imaging); PWI (Perfusion-Weighted Imaging); ASL (Arterial Spin Labeling); DSC (Dynamic Susceptibility Contrast); DCE (Dynamic Contrast-Enhanced); PET (Positron Emission Tomography); FDG (Fluorodeoxyglucose); MRS (Magnetic Resonance Spectroscopy).
Structural imaging: Despite the development of new imaging modalities over the past several decades, conventional MRI and CT that evaluate structural components prevail as the most common imaging modalities for high grade gliomas. The current standard parameters to monitor tumor progression are with contrast enhancement on T1 MRI and the extent of T2 hyperintense areas on FLAIR MRI [37]. However, as mentioned previously, enlargement of contrast enhancement or widening of the hyperintense areas can be nonspecific [16]. Furthermore, since high grade gliomas affect and invade brain tissue at the molecular level, the borders of the tumors cannot be well visualized with conventional MRI [11].
Functional imaging, physiologic imaging: Functional imaging has become more heavily utilized in high grade glioma patients. Several of these advanced imaging techniques that provide physiologic information such as Diffusion Weighted Imaging (DWI) MRI and Perfusion Weighted Imaging (PWI) MRI are already utilized in clinical practice [11].
DWI MRI
DWI MRI provides physiologic data by assessing the microscopic motion of water molecules in the brain parenchyma in order to evaluate the tumor microenvironment [37,38]. In areas that water diffusion is focally restricted, DWI MRI shows as a bright image signal [39]. Through mathematical subtraction of unwanted T2 effects, it provides the Apparent Diffusion Coefficient (ADC) map, on which dark signal corresponds to areas of restriction [40]. The ADC has been found to correlate with the cellularity of tumors [41]. High grade gliomas are expected to have low Apparent Diffusion Coefficient (ADC) values because of their high cellularity and progressive tumor often demonstrates lower mean ADC values than normal brain tissue and necrotic debris [38,42]. Previous studies have shown differences in the ADC values of PsP and PD, but with different ADC thresholds [42–44].
PWI MRI
On the other hand, PWI provides hemodynamic characteristics of brain lesions such as cerebral blood flow, cerebral blood volume, permeability estimates, and contrast time peak in order to evaluate the tumor microvasculature [38]. There are three major perfusion MRI techniques, Arterial Spin Labeling (ASL), Dynamic Susceptibility Contrast (DSC), and Dynamic Contrast- Enhanced (DCE) MRI. Unlike ASL, both DSC and DCE are acquired during IV contrast administration [45]. ASL-MRI involves magnetically labeled arterial blood water protons within circulating blood before entering the brain instead of contrast for perfusion characterization [46]. This technique is much less frequently used compared to the other perfusion modalities [46]. DSC MRI measures alterations in T2/T2* (gradient echo) signal intensity during the first phase of an IV injection of gadolinium contrast [47]. It is the most commonly used functional imaging technique due to its relatively short imaging time and the widespread availability of the post-processing software [48]. It obtains data that gives several parameters that can be quantified and compared such as relative cerebral blood volume (rCBV) [48]. DCE MRI, or permeability imaging, allows for the measurement of vascular permeability pharmacokinetics and utilizes T1 relaxation of contrast with fast imaging acquisition [37,49]. It is often a useful adjunct for lesions that have indeterminate rCBV values, susceptibility artifact from blood products, or calcification that prevents DSC quantifications [50]. Due to the vascular hyperpermeability and BBB disruption in high grade gliomas, the IV contrast can leak from intravascular compartments to the extravascular space, subsequently leading to an increase in T1 signal intensity from the paramagnetic effect [51]. However, DCE quantification requires challenging pharmacokinetic modeling in which there are a variety of methods with accepted standard [52]. Thus, it is less commonly used in daily practice than DSC MRI. The limitations of PWI include the effects of BBB disruption, extravascular contrast leakage, and different post-processing software tools on the accuracy of rCBV calculations [53-55].
Molecular imaging
Advanced MRI, PET, and SPECT can be modified and used for both physiologic and molecular imaging. Physiologic imaging evaluates differences in perfusion, fluid diffusion, and environmental conditions to differentiate between healthy and malignant brain tissue [11]. Molecular imaging modalities visualize specific proteins, receptors, or metabolites associated with a targeted pathology, tissue, or cell population [11].
FDG PET
Imaging techniques can also be harnessed and modified to provide molecular imaging data. One example is Fluorodeoxyglucose (FDG) PET, which is the most widely used molecular agent for PET [56]. PET measures metabolic uptake and FDG PET uses FDG, which is a marker for glucose metabolism that is transported across the BBB to CNS tissue, as a metabolic tracer to elucidate different tumor metabolic properties [38]. When using FDG PET, high-grade tumors like glioblastoma generally demonstrate increased FDG metabolite uptake due to their elevated metabolic state, while radiation necrosis has low FDG uptake [38].
Amino acid PET
Amino acid PET is another type of PET technique that utilizes tracers with greater turnover in metabolically active tumors that demonstrate lower background uptake in normal brain than FDG [57]. These tracers include 18-F-L-Fluoro- DOPA ([18F]- DOPA), 3'-deoxy-3'[(18)F]-fluorothymidine ([18F]-FLT), 18Ffluoro- ethyl-tyrosine ([18F]-FET), and L-[methyl-11C]-methionine ([11C]-methionine) [58].
Antibody-based PET
Antibody-based PET is also a technique that is being more frequently used in neuro-oncology. It uses antibodies or antibody derivatives that bind to prognostically or therapeutically relevant receptors or proteins expressed in tumor and immune cells such as PD-1, PD-L1, CTLA-4, and EGFR [11]. These macromolecules access high grade gliomas and CNS metastases through disrupted areas of the BBB, but cannot access CNS tissue well when the BBB is intact such as nonenhancing regions of high grade gliomas or low-grade gliomas [59].
Reporter transgene imaging
Another molecular imaging approach has been the use of an MRI, PET, or SPECT molecular reporter probe. This probe interacts with the product of a reporter gene that is introduced into desired cells to express a specific protein that is not normally expressed in non-target tissues [60]. The probe accumulates in the target tissue and is able to be imaged [60]. This approach is also known as reporter transgene imaging and it is useful in longitudinal imaging of cell viability and trafficking following introduction of the reporter gene [61]. Like antibody-based PET, these reporter molecules must be able to cross the BBB in order to detect target cells throughout CNS tissues [61].
MRS
Lastly, Magnetic Resonance Spectroscopy (MRS) is a molecular imaging technique that is being used in patients with suspicion of a high grade tumor. It examines the distribution of chemical metabolites within a designated volume of tissue [62]. Two main MRS techniques are Single Voxel Spectroscopy (SVS) and Multi- Voxel Spectroscopy (MVS) [63]. SVS has a quicker turnaround but cannot capture spatial heterogeneity whereas MVS requires more complex preparation and post-processing, as well as longer wait times [63]. Although protons are the metabolites most commonly used for MRS, other nuclei such as metal ions and phosphorus have also been utilized [62,64].
Clinical studies incorporating functional imaging techniques for immunotherapy
While the above imaging modalities have all been utilized in high grade glioma patients, few studies have explored how these modalities can be used to monitor response to immunotherapy.
Structural imaging modalities have been the standard to assess treatment response, but with the development of more advanced imaging techniques, functional imaging has been a particular area of interest [11]. Immunotherapy as a field has become quite vast and now encompasses a variety of modalities: checkpoint inhibitors, viral, cell-based, and vaccine immunotherapy.
Checkpoint inhibitor immunotherapy
Checkpoint inhibitor immunotherapy is used to overcome immunosuppressive cell-cell signaling mechanisms between cancer cells and effector immune cells such as cytotoxic T lymphocytes [65]. Monoclonal antibodies that bind to Programmed Death-1 (PD-1), Programmed Death-Ligand 1 (PDL1), and Cytotoxic T Lymphocyte-Associated protein 4 (CTLA-4) have proven successful at generating immune responses in groups of patients with various solid tumor malignancies such as non-small cell lung cancer, renal cell carcinoma, and melanoma [7]. Thus far, the only completed randomized phase III trial assessing the impact of nivolumab (PD-1 checkpoint immunotherapy) on glioblastoma patients showed no survival compared with those who received bevacizumab [66]. There are various ongoing clinical trials that are assessing the efficacy of other checkpoint inhibitor immunotherapies in glioblastoma patients [67,68].
Viral-based immunotherapy
Viral-based immunotherapy uses live, immunogenic viruses that replicate in tumor cells to stimulate immune responses within the tumor microenvironment [69]. Oncolytic virotherapy is based on harnessing replicating viruses that selectively kill only the infected cancer cells rather than normal cells by directly lysing the host tumor cells, thereby releasing additional tumor antigens to enhance the immune response [69]. In glioblastoma, oncolytic virotherapies are able to change the TME to favor an antitumor response by reprogramming tumor-associated macrophage polarization [70].
Cell-based immunotherapy
In cell-based immunotherapy, precursor cells from the innate or adaptive immune system are collected from healthy donors or patients, activated or transformed ex vivo, and infused into the patient as immune cells that recognize and destroy specific types of antigen-presenting tumor cells without needing priming via antigen presenting cells [71]. These cell-based immunotherapies are varied and include T-cell receptor-transduced T-cells, lymphokine-activated natural killer cells, and Chimeric Antigen Receptor (CAR) T-cells [72,73].
Vaccine immunotherapy
Lastly, vaccine immunotherapy stimulates immune responses by presentation of tumor cell antigens in antigen presenting cells to T cells to lead to T cell clonal expansion, circulation, and destruction of antigen-bearing tumor cells [74]. These vaccine strategies employ various techniques that include but are not limited to tumor cell-based, peptide-based, and dendritic cellbased techniques [75,76].
Physiologic imaging in glioblastoma patients
While advanced functional imaging techniques are more commonly used in other solid tumor immunotherapy trials, they are becoming increasingly used to monitor glioblastoma patient response to different immunotherapeutics (Table 1). Physiologic imaging such as DWI MRIs have been used in glioblastoma clinical trials using checkpoint inhibitor therapy.
| Imaging Technique Category | Imaging Modalities | Type(s) of Immunotherapy Received | Number of patients | Reference |
|---|---|---|---|---|
| Physiologic | ||||
| DWI* MRI | Checkpoint inhibitor | 10 glioblastomas | [77] | |
| Multiparametric MRI | Checkpoint inhibitor | 19 glioblastomas | [78] | |
| Multiparametric MRI | Cell-based | 10 glioblastomas | [79] | |
| PWI* MRI, DWI* MRI | Vaccine | 8 glioblastomas | [80] | |
| PWI MRI | Vaccine | 22 glioblastomas | [81] | |
| Molecular | ||||
| [124I]-FIAU PET* reporter probe | Viral | 5 glioblastomas | [60] | |
| [124I]-FIAU SPECT* | Viral | 8 glioblastomas | [83] | |
| PET reporter gene | Cell-based | 1 glioblastoma | [85] | |
| PET reporter gene | Cell-based | 7 glioblastomas | [61] | |
| [11C]-methionine uptake PET | Vaccine | 14 glioblastomas | [87] | |
| 18F-FET PET* | Vaccine | 3 glioblastomas | [90] | |
| [18F]-CFA PET* reporter probe | Vaccine +/-checkpoint inhibitor | 3 glioblastomas | [92] | |
| Radiomics | ||||
| Features extracted from conventional, PWI* or DKI* MRI Best performance with AdaBoost*, RUSBoost*, and SVM-rbf* trained mainly on features extracted from T1pc* and CBV* maps |
Vaccine | 29 glioblastomas | [116] | |
Abbreviations: *DWI: Diffusion Weighted Imaging; PWI: Perfusion- Weighted Imaging; FIAU PET- Furanosyl-5-Iodo-Uracil Positron Emission Tomography; FIAU SPECT: Furanosyl-5-Iodo-Uracil Single Photon Emission Computed Tomography; DKI: Diffusion-Kurtosis Imaging; AdaBoost-adaptive boosting algorithm; RUSBoost: Random Undersampling Boosting Algorithm; SVM-rbf: Support Vector Machine Radial Basis Function; T1pc: T1 Postcontrast; CBV: Cerebral Blood Volume
Table 1: Imaging techniques and methodologies used for immunotherapy treatment response in high grade glioma patients.
One study by Qin et al. used DWI MRIs to evaluate response to checkpoint inhibitor in ten patients with recurrent glioblastoma [77]. These patients were treated with anti-CTLA-4 and/or anti- PD-1 CPIs and the researchers showed that intermediate ADC (IADC) DWI volume changes within FLAIR regions of high cellularity was correlated with immunotherapy response [77]. Five out of the ten patients were deemed to have a therapeutic response to the CPIs and they were identified by the IADC changes as faster when compared to conventional MRI (median time of 93 vs. 121 days after treatment) [77]. The study also showed that therapeutic outcome defined as time to progression, was more strongly correlated with the IADC volumes than with Gd-contrast enhancement, FLAIR, or 2D RANO measurements [77]. The authors acknowledge that although these preliminary findings are promising, they require further exploration in larger human cohorts as well as in animal models to better understand the relationship between IADC volumes and immunotherapy treatment response [77]. Furthermore, although there was an improvement in treatment response detection in using IADC over conventional MRI, it is important to identify response even earlier than the three months to switch non-responding patients more quickly to an alternative treatment.
Song et al. published a small retrospective study utilizing relative ADC (rADC) changes to determine six month progression free survival in a group of nineteen recurrent glioblastoma patients receiving anti-PD1 immunotherapy (either nivolumab or pembrolizumab) [78]. Out of the nineteen patients that met inclusion criteria and had sufficient follow-up, seven were determined to have treatment response whereas twelve were determined to have PD after six months using the mRANO criteria [78]. They compared quantitative values of imaging biomarkers such as rADC, Ktrans, Vp, Ve, and rCBV in preand post-ICI MRIs and only changes in rADC values were indicative of treatment response [78]. Six out of seven patients with treatment response had interval increased rADC, whereas eleven out of twelve patients with no treatment response had interval decreased rADC (p=0.001) [78]. Although this study showed that physiologic imaging data could be used to effectively differentiate treatment responders from nonresponders, rADC values were collected after six months and not earlier.
Physiologic imaging has not been frequently used for cell-based immunotherapy. One study used multiparametric MRI parameters obtained from DTI, DSC, and 3-D echo planar spectroscopic imaging (3D-EPSI) to monitor treatment response to EGFRvIII targeted CAR-T cell therapy in ten recurrent glioblastoma patients [79]. Using a set of obtained parameters, they computed progression probabilities using logistic regression models and were able to objectively characterize each lesion as PsP, PD, or treatment response at each individual time point (one, two, and three months from treatment initiation) [79-116].
Physiologic imaging has also been used as a treatment response modality in patients receiving vaccine immunotherapy. In a pilot study, eight recurrent glioblastoma patients were treated with a dendritic cell vaccine and the researchers used both PWI and DWI MRIs to differentiate between immunotherapy-related PsP versus PD [80]. They showed that the highest rCBV and lowest minimal ADC (rADCmin) in the contrast-enhancing regions were associated with PD, but they did not provide any relationship with treatment response [80]. Cuccarini et al. used DWI and DSC MRIs to evaluate response to a specific dendritic cell vaccine in twenty-two newly diagnosed glioblastoma patients [81]. Similar to the song group, they were able to use rADC to predict response to immunotherapy and survival [81]. They showed that patients who were DC vaccine treatment responders, with elevated NK cells post-treatment, had a significant decrease in rADCmin when looking at pre and posttreatment values, compared to the non-responders [81]. Furthermore, they showed that when comparing imaging from two months prior to PsP or PD, ΔrCBVmax ≥ 0.47 distinguished TTP from PsP with a sensitivity of 67% and specificity of 75% (p=0.004) [81]. Previously, an earlier study looking at a group of patients treated with an autologous irradiated tumor cell vaccine had not shown any relationship between rCBV values with treatment or survival time [82]. Cuccarini’s group, however, showed that longitudinal rCBV changes could be useful in distinguishing PsP from PD [81].
Molecular imaging in glioblastoma patients
PET and SPECT imaging techniques have been used for glioblastoma patients being treated with gene therapy and oncolytic viruses. Jacobs et al. used PET imaging to monitor the response of five patients with recurrent glioblastoma to the transduction of a genetically modified herpes simplex virus type-1 thymidine kinase (HSV-1-tk) gene with subsequent prodrug activation by ganciclovir [60]. The HSV-1-tk acts as a safety gene, such that when exposed to the chemotherapeutic ganciclovir, the virus will undergo programmed cell death [60]. The PET reporter probe, I-124-labelled 2’-fluoro-2’-deoxy-1-Darabino- furanosyl-5-iodo-uracil ([124I]-FIAU), was a specific marker substrate for gene expression of the transduced HSV-1-tk gene in order to identify the location, magnitude, and extent of the vector-mediated gene expression [60]. The gene therapy proved to be safe in all five patients [60]. However, the probe only accumulated in one out of five patients and did not provide any therapeutic benefit in any of the patients [60]. Another group used SPECT imaging to monitor response to oncolytic viral therapy in eight glioblastoma patients [83]. Similar to the Jacobs group, they used a modified HSV-1-tk gene and a radiolabeled thymidine analog, [123I]-FIAU, to detect expression of the gene and the viral distribution [83]. They were unable to show accumulation of the reporter probe in any of the eight treated patients [83]. Overall, for viral-based immunotherapies, it is unclear how effective molecular imaging techniques are to monitor treatment response. Significant progress needs to be made in these techniques in differentiating PsP and PD.
Molecular imaging techniques have also shown some promise in assessing treatment response in glioblastoma patients receiving cell-based immunotherapies. This approach includes using molecular imaging of immune or inflammatory cells to identify patients demonstrating effective immune cell trafficking or activation in the tumor in a manner that is more effective, less invasive, or quicker than relying on conventional imaging or biopsy to determine PD. Typically, cells will be labeled with a contrast agent for the respective imaging machine such as MRI, PET, or SPECT prior to injection into the patient [84]. The injected cells are then monitored to determine the cell localization and trafficking in the target tumor region [84].
Yaghoubi et al. developed a specific PET imaging system to detect the genetically engineered autologous CD8+ CTLs injected into a glioblastoma patient, difficult by conventional imaging methods alone [85]. In a case report, a single glioblastoma patient was enrolled in a clinical trial of adoptive T cell therapy with genetically engineered CTLs expressing the PET imaging reporter gene, the HSV-1-tk gene, and the CAR IL-13 zetakine gene [85]. The IL-13 zetakine allowed the engineered CTLs to specifically target and kill residual glioblastoma cancer cells and the HSV-1-tk enzyme monophosphorylates [18F]-FHBG so that PET can be used in living organisms to image cells that express the PET reporter gene [85]. The [18F]-FHBG scans showed significantly higher signal after the infusion of the engineered CTLs within the tumor lesions of the brain and they were successful in showing that this reporter probe can penetrate the BBB [86]. This led to a larger clinical trial (NCT01082926) that has been completed, but the efficacy of the cell-based therapy or the use of PET for treatment response detection have not yet been published. Nevertheless, Keu et al. demonstrated that the PET imaging system developed by Yaghoubi using [18F]-FHBG could be used in longitudinal monitoring of the proliferation, trafficking, and survival of adoptively transferred genetically engineered CTLs [61]. In seven glioblastoma patients in this study, the therapy was safe, and the imaging technique was noted to be sufficient to properly locate and track the infused CTLs [61]. The uptake of the tracer was increased in all brain lesions after infusion of the CTLs compared to the pre-treatment PET, but the volume of distribution and pattern of increase in PET signal intensity varied between patients [61]. Due to the small number of patients enrolled in this study, it was unable to link changes in [18F]-FHBG signal before and after adoptive T cell infusions to patient outcome and survival [61]. Given that [18F]-FHBG was the first PET reporter probe to receive investigational new drug approval from the FDA (IND #61,8880) and the enormous cost of ACT therapy (approximately $200,000/individual), it has been difficult to get enough patients to help further refine and understand this imaging technique [61]. These studies demonstrate the importance of differentiating between specific immune cell trafficking and localization within glioblastoma lesions and non-specific pooling of PET agents to assess the efficacy of cell-based immunotherapy through reporter gene physiologic imaging techniques.
Vaccine immunotherapy for glioblastoma has also utilized molecular imaging techniques to determine treatment response. Chiba et al. used PET molecular imaging to define parametric response maps in fourteen recurrent glioblastoma patients who received the WT1 peptide vaccination [87]. WT1 gene products have been shown to be overexpressed in glioblastoma, thus making the WT1 antigen a viable target for immunotherapy [88,89]. The authors used 11C-methionine uptake PET prior to treatment and twelve weeks post-treatment. Voxel-wise PET analysis in MRI contrast-enhancing areas of the tumor and showed that a 5% increase of 11C-methionine was a sufficient threshold to differentiate treatment non-responders from responders [87]. In contrast, conventional MRI was not helpful in identifying those who responded to treatment [87]. In another study, the authors examined five patients who received dendritic cell vaccine treatment as well as 18F-fluoroethyltyrosine (18F-FET) PET imaging and conventional MRI [90]. In three patients, 18F-FET PET was able to confirm PD using the RANO criteria for conventional MRI [90]. In three patients, they were able to correctly determine that a patient was exhibiting PsP with 18F-FET PET whereas conventional MRI incorrectly deemed it as PD [90]. The 18F-FET PET imaging findings were confirmed by histopathology and RANO criteria [90]. These specific PET images were obtained within six weeks of initiating vaccine immunotherapy, thus potentially providing accurate treatment response information much faster than conventional imaging techniques [90]. Furthermore, 18F-FET PET imaging provided more accurate diagnosis of PD over conventional MRI alone after standard therapy in a larger glioblastoma patient study [91]. Although promising, much larger studies are needed to confirm its potential.
In another study, three glioblastoma patients were given a PET probe for deoxycytidine kinase (dCK), 2-chloro-2′-deoxy-2′- [18F]fluoro-9-b-D-arabinofuranosyl-adenine ([18F]-CFA), and imaged by PET prior to and following treatment with a DC vaccine and either pembrolizumab or bevacizumab [92]. These patients were also imaged by PWI and DWI MRI and the researchers used the imaging data combined with the information provided by the PET scans to create an immunotherapeutic response index (ITRI) [92]. This index determined through these imaging modalities was correlated with response to the combination immunotherapy and thus were able to noninvasively localize and quantify the immune responses induced by immunotherapy [92]. The authors suggested that this novel index and combination of imaging modalities could standardize how immunity and immune response to these therapies are measured and may serve as a reproducible predictive biomarker of survival and treatment response in glioblastoma patients [92].
While there is a lot of promise in the imaging field to detect treatment response in glioblastoma patients receiving immunotherapy, certain limitations exist. Many of the studies are retrospective with small patient numbers. They utilize varying imaging protocols and techniques, which limits the ability to use them in routine clinical practice and makes comparison difficult. Thus, standardization is essential. Future studies will require a much larger number of patients to provide more definitive evidence of their efficacy and to demonstrate which modalities have the most reliable metrics for monitoring immunotherapy response.
Radiomics
Given the current limitations of even the most cutting edge of advanced imaging modalities, integration of imaging with radiomics is increasingly attractive. Radiomics is a biomedical field that involves the computational and statistical extraction of large amounts of advanced quantitative data from a variety of imaging modalities such as MRI, CT, SPECT, PET, and others, via machine learning methods in order to develop predictive models that aim to enable a more personalized medical approach [93]. This field is producing biomarkers to provide key insights in the diagnosis, classification, and therapeutic management of various solid tumors as well as synergy with other data such as molecular biomarkers and patient characteristics to facilitate diagnostic and treatment decision support models [94].
There are many iterations and variations of radiomics analysis pipelines based on the imaging modalities and statistical methods used. However, several common overarching steps are involved: preprocessing and image acquisition for development of the radiomics model; tumor labeling/segmentation; identification of relevant features that may relate to the molecular properties of the tumor; and statistical modeling correlating the radiomic analysis information with a diagnosis or clinical outcome [95]. After the images are initially acquired from their respective modalities, the lesions are identified and image segmentation occurs, which can be an automated or manual process, as well as 3D reconstruction of the Region Of Interest (ROI) [95]. Then, the next important step is feature extraction and classification [95]. Radiomics features are defined as characteristics of the imaging modalities that are too complex for a human to determine or appreciate independently and are the basis for the various radiomics analyses that are performed [96]. These radiomics features include quantitative descriptions of the shape, volume, size, intensity, and texture of the region of interest (ROI) [97]. They are also divided into texture (first-order statistics) and histogram-based (second-order statistics) features [97]. Feature extraction produces several numerical values and can be analyzed using advanced statistical and machine learning methods [98]. These machine learning methods can be either supervised or unsupervised, and can include random forest, support vector machine, cluster analysis, convolutional neural network, and deep learning neural network [98].
Tissue biopsies can fail to characterize the entirety of the tumor due to heterogeneity seen in high grade glioma tumors [99]. Radiomics, however, may become an important tool by taking the whole tumor region into account for better characterization [100].
Radiomic studies for glioblastoma diagnosis, prognosis, and progression
One aspect of radiomics is the incorporation of multiple image features to create a noninvasive molecular signature that can characterize glioblastoma and predict survival. Gevaert et al. showed that the use of quantitative imaging values, such as irregularity of border edges in a certain ROI, correlated significantly with overall survival and characterization of glioblastoma with regard to molecular subtypes [101]. Moreover, they were able to generate sets of imaging features that correlated with patients’ underlying gene expressions [101].
Through the creation of a “radiogenomic map,” imaging characteristics could be used to derive genomic and clinical data [101]. Yang et al. ran a similar analysis after manual segmentation of tumor from postcontrast T1 and T2-FLAIR images to extract a wide variety of imaging texture features [102]. Using computational methods to pick subsets of the features, the authors generated ROC curves for prediction of molecular subtype and twelve-month survival [102]. The area under curve values ranged from 0.70-0.82 for prediction of molecular subtype and was 0.69 for prediction of twelve-month survival [102].
Lee et al. also created a predictive model for identification of molecular subtype and twelve-month survival estimates [103]. In this model, the authors conducted a spatial point pattern analysis on T1 post-contrast and T2-FLAIR images [103]. The prediction of twelve-month-survival based on these analyses yielded a sensitivity and specificity of 0.86 and 0.64, respectively [103]. The accuracy of the ROC curves in predicting molecular subtype of the glioblastomas ranged from 0.7-0.93 [103]. Jamshidi et al. constructed radiogenomic maps that also incorporated mRNA expression and copy number variations in the genetic analysis [104]. Six imaging features (contrast enhancement, necrosis, contrast-to-necrosis ratio, infiltrative versus edematous T2 abnormalities, mass effect, and subventricular zone involvement) were used to create a model that was then correlated to mRNA expression and copy number variation [104]. This radiogenomic map identified specific genes that were then correlated to tumor aspects including aggressiveness, edema, and mass effect [104].
Hsu et al. showed that the immune component of the tumor microenvironment could be characterized based on radiomics analysis [105]. After collection of RNAseq data, the authors clustered the immunophenotypes of the 154 glioblastoma samples into five categories [105]. T1 post-contrast and ADC values were then collected to extract features which could then be used to correlate one of the five immunophenotypic subsets [105]. Accuracies for the T1 contrast model ranged from 0.72-0.88 in predicting the immunophenotypic breakdown while accuracies for the ADC model ranged from 0.61-0.79 [105]. After patients were classified based on these radiomics models, the authors showed that patients who displayed a more favorable immunophenotypic subtype on imaging had a better overall survival (OS) [105]. Overall, these studies show the capacity of radiomics to provide a non-invasive method to gain insight to the tumor phenotype and predict patient survival.
Radiomic studies for glioblastoma adjuvant treatment response
There have also been studies that focus on determining treatment response in glioblastoma patients using radiomics analyses. Artzi et al. utilized unsupervised clustering analysis of conventional, DSC, DWI, DSC, and DCE features to classify FLAIR abnormalities in fourteen glioblastoma patients receiving bevacizumab therapy [106]. A total of 40 MRIs were analyzed to create a radiomics model to differentiate PsP from PD [106]. This model was correlated with patient outcome including PFS at weeks eight and sixteen post-therapy [106].
Chang et al. also harnessed radiomics to develop a predictive model for bevacizumab therapy response in glioblastoma patients [107]. The study included 126 patients with recurrent glioblastoma, 84 patients who were placed in the training cohort and 42 patients in a testing cohort for the radiomics model. Preand post-therapy DWI and conventional MRI features were inputted into a random forest algorithm to create a model that predicted OS with a hazard ratio of 5.10 (p<0.001) in the training cohort and 3.64 in the testing cohort (p<0.005) [107]. Kickingereder et al. used a radiomic model and implemented a supervised principal component analysis to generate a prediction model for stratifying treatment outcome to bevacizumab for both PFS and OS in patients with 172 recurrent glioblastoma patients [108]. Their radiomics analysis used conventional MRI sequences and stratified patients into a low or high risk group for PFS (HR=1.60; p=0.017) and OS (HR=2.14; p<0.001) and was successfully validated for patients in their validation cohort for both PFS (HR=1.85, p=0.03) and OS (HR=2.60, p=0.001) [108]. These studies provide promising radiomic biomarkers that could more effectively determine treatment response in glioblastoma patients than conventional imaging or advanced imaging techniques alone.
Radiomic studies for immunotherapy response
While the application of radiomics to monitor response to immunotherapy response is a natural extension of the prior studies, there is a paucity of data in the literature for glioblastoma. There have been investigations into the use of radiomics for monitoring immunotherapy response in patients with other solid tumors. Sun et al. performed a retrospective multicohort study in which four independent cohorts of patients with advanced solid tumors were included [109]. RNAsequencing data derived from tumor biopsies and pretreatment images from these cohorts were used to develop and validate a radiomics biomarker predictive of immunotherapy response and CD8 T cell tumor infiltration [109]. They developed a Radiomics Signature (RS) of CD8 T cell count comprised of eight radiomics features based on the 135 patients enrolled in the Molecular Screening for Cancer Treatment Optimization (MOSCATO) trial, which was validated with an independent cohort of 119 patients from The Cancer Genome Atlas (area under the curve (AUC)=0.67, p=0.0019) [109]. This signature was further validated in two other cohorts [109]. The first independent cohort included 100 patients with tumors classified as either immune-desert or immune-inflamed (AUC=0.76, p<0.0001) with either poor or dense CTL infiltration [109]. The second independent cohort of patients treated with ICIs demonstrated that the signature was associated with clinical outcomes; a high baseline radiomics score correlated with an objective response instead of stable disease or progression (p=0.025) and an improved overall survival (p=0.0022) [109]. One of the eight radiomics features in this signature was a technical variable, the peak kilovoltage, a finding that the authors noted highlighted the importance of image acquisition parameters [109]. This study is of great interest since it is the first immunotherapy study to link imaging, tumor phenotype, and clinical responses to treatment. A later study validated this radiomics signature in 68 patients with 139 metastatic solid tumors who received pembrolizumab and stereotactic radiosurgery (SRS) [110]. The RS was treated as a dichotomous variable, with the bottom 25% or lower defined as low-RS [110]. Patients with low-RS were significantly less likely to be SRS responders (p=0.012), and had worse PFS (median: 2.76 vs. 3.38 mo., p=0.01) and OS (median: 6.21 vs. 13.3 mo, p=0.005) [110]. Patients with high-RS correlated with transcriptional activation of adaptive and innate immune signatures in biopsies from irradiated tumors, which could suggest those that are SRS and treatment responders [110].
Another radiomics study that investigated response prediction to immunotherapy was by Trebeschi et al. in 203 patients with advanced melanoma and non-small-cell lung cancer (NSCLC) undergoing anti-PD1 ICI therapy [111]. In this study, pretreatment CTs of patients with 1055 primary and metastatic lesions were used to build a machine learning model to predict treatment response at the lesion level [111]. The model proved to be significant for both melanoma nodal metastases (AUC=0.64, p=0.05) and NSCLC pulmonary and nodal metastases (AUC=0.83 and 0.78, p<0.001) with better predictive performance for NSLC metastases [111]. By combining the lesion-specific data on the patient level, they were able to predict immunotherapy response with an AUC of up to 0.76 (p<0.001) for both cancer types, with a one-year survival difference of 24% (p=0.02) [111]. The authors also performed biological validation of their radiomics biomarker using an independent cohort of 262 NSCLC patients with pre-operative CT and gene expression data [111]. The top gene sets most strongly correlated with their biomarker were those involved in cell cycle progression and mitosis [111].
Basler et al. also demonstrated the predictive potential of PET/CT-based radiomics in a retrospective cohort of 112 metastatic melanoma patients being treated with ICIs [112]. They used pretreatment CT and PET for 716 baseline metastases, tumor lesion volume, and the routine blood markers LDH and S100 to help differentiate PsP from PD [112]. Using this information, they developed seven different model classes, including models that used each modality separately (blood biomarkers, lesion volume, and radiomics) as well as various combinations of the modalities such as blood and lesion volume or blood biomarkers and radiomics [112]. The best performing model was the combined blood biomarkers and radiomics model excluding the volume-related features (AUC=0.82) [112]. This study lends support to the idea that a multi-modal approach will be best to provide early determination of immunotherapy treatment responders [112].
Another aspect of immunotherapy treatment is the development of Immune-Related Adverse Events (irAEs) in a subgroup of patients. These irAEs are diverse and have important clinical implications with the most common being dermatitis, endocrinopathies, enterocolitis, hepatitis, pneumonitis, transaminitis, and uveitis [113]. Being able to quickly identify patients that will develop severe irAEs is therefore paramount. Colen et al. published a proof of concept study demonstrating the potential of radiomics to predict patients who are at risk for developing immunotherapy-induced pneumonitis [114]. Although it is not as common as some of the other irAEs, it is of particular concern given its life threatening nature [115]. This retrospective study involved a group of patients with advanced cancer who were enrolled in early phase immunotherapy clinical trials that had been treated with at least one immunotherapeutic agent [114]. They used pretreatment CTs from two patients who developed pneumonitis and as a control, a random thirty patients who did not [114]. Six volumes of interest for each patient were delineated with a total of 1860 features extracted per patient [114]. They used a Maximum Relevance Minimum Redundancy (MRMR) feature selection method to identify features and rank them based on their relevance to the outcome (immunotherapy-induced pneumonitis) and redundancy [114]. Immunotherapy-induced pneumonitis was predicted using an unsupervised anomaly detection algorithm [114]. They showed that the most predictive features were skewness (measurement of histogram symmetry) and angular variance of sum of squares (measure of dispersion of the voxel intensity distribution), with 100% accuracy (p=0.00033) [114].
Radiomics for immunotherapy response in glioblastoma patients
Currently, there is only one study in the literature regarding the use of radiomics to track response to immunotherapy for glioblastoma. Ion-Margineanu et al. examined whether analysis of MRI, PWI, and diffusion kurtosis MRI (DKI) images using a radiomics pipeline could distinguish progressive disease or treatment response when treated with dendritic cell immunotherapy [116]. By acquiring imaging texture features and histograms of MRI intensity values in both a manual or semimanually outlined ROI model, the authors created a pipeline to assess treatment response [116]. The model was scored based on a Balanced Accuracy Rate (BAR) consisting of the average of the sensitivity and specificity [116]. Features extracted from T1 postcontrast MRI and CBV maps in manually delineated ROIs trained with the adaptive boosting algorithm (AdaBoost) algorithm yielded the highest BAR value of 0.956 [116]. For further automation, algorithms created from a combination of T1 post contrast features and cerebral blood volume readings from semi-manually delineated ROIs yielded BAR values of 0.947 and 0.932 when using the random undersampling boosting algorithm (RUSBoost), or support vector machine radial basis function (SVM-rbf), machine learning algorithms respectively [116]. The findings suggest potential use of radiomics pipelines to assess therapeutic response to immunotherapy in glioblastoma.
Despite radiomics being an extremely promising field to accelerate personalized medicine for glioblastoma patients, challenges and limitations exist. One of its biggest challenges is the field’s data reproducibility. Given the novelty as well as the technical complexity of this methodology, there is currently significant variability in segmentation techniques and feature extraction algorithms that can adversely impact the sensitivity of the radiomics features [117]. Another important obstacle is data sharing, a common challenge in all biomedical research, as it must overcome administrative, regulatory, cultural, and personal difficulties and comply with the Health Insurance Portability and Accountability Act (HIPAA) [118,119]. These past radiomics studies involve small sample sizes that make it difficult to draw significant conclusions and generalizations. Data sharing problems need to be addressed to further advance this field.
High grade glioma patients face a grim prognosis with often ineffective treatments and potentially significant side effects. They must undergo a complex, and often, invasive clinical workup to assess response to the treatment provided. As immunotherapy is being investigated as a treatment for high grade gliomas, better tools to assess real-time response to immunotherapy are needed. Pseudoprogression mimicking true progression of disease is an issue that is especially important. Patient survival time is extremely short and determining treatment response earlier even by a few months can make a significant difference. Innovations in advanced imaging techniques and modalities facilitated more effective and quicker evaluations of the treatment efficacy. Significant hurdles remain to incorporating effective advanced imaging strategies into routine clinical practice. Relatively low incidence of high grade gliomas, the high cost of performing these studies, and the low number of patients enrolled in immunotherapy clinical trials pose a huge challenge in accruing adequate numbers of patients needed to train and test machine learning algorithms to differentiate immunotherapy responders from non-responders. A report showed that only one in three of neuro-oncological studies published after 2010 used the RANO criteria and with the recent development of other criteria, a standardized response criteria and terminology across the neuro-oncology field are needed. With validated guidelines to compare these multi-modal advanced imaging techniques, their use will become more routine to detect treatment response.
Radiomics is a powerful tool that can help resolve some of the challenges found in advanced imaging to provide important information on diagnosis, prognosis, progression, and treatment response in high grade glioma patients. However, it has its limitations and challenges that must be overcome. It has the advantage of noninvasively providing information about the whole tumor environment but, it is operator-dependent. Radiomics will be one important tool in a toolbox of various techniques such as patient characteristics, genomics, metabolomics, and transcriptomics that in the future will together provide a more personalized medical approach.
Citation: Kim T, Mathios D, Pan C, Srivastava S, Lim M (2021) Advances in Imaging Techniques for Disease Monitoring of High Grade Gliomas during Immunotherapy. J Clin Trials. 11:457.
Received: 03-Feb-2021 Accepted: 17-Feb-2021 Published: 24-Feb-2021 , DOI: 10.35248/2167-0870.21.11.457
Copyright: © 2021 Kim T, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.