Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research Article - (2015) Volume 4, Issue 2
The QSAR studies were performed on a series of benzimidazole derivatives to find out the structural requirements of their antimicrobial activities. The QSAR studies have been carried out using electronic, thermodynamic, spatial and structural descriptors along with a few structural parameters. The best QSAR (S.aureus) model having r2= 0.7886 and q2 = 0.7269 was developed by principle component regression. The results derived may be useful in further designing of more potent antimicrobial agents in series. Analysis of 2D-QSAR models provides details on the fine relationship linking structure and activity and offers clues for structural modifications that can improve the activity.
<Keywords: benzimidazole, 2D-QSAR, VLife MDS, antimicrobial activity, principle component regression
Over the past few decades, the problems posed by multi-drug resistant microorganisms have reached an alarming level in many countries around the world [1]. This has led to a serious challenge to the medical community; hence, the development of new effective antimicrobial agents is an important goal. In pursuit of this goal, our research efforts are focused on the development of novel structural moieties with promising antimicrobial properties [2,3]. Bacterial infections are still a major cause of death in the developing world. Another problem is resistant bacteria from hospitals, which cause ‘community-acquired’ infections, difficult to treat, particularly in immuno-compromised patients. These resistant bacteria have also acquired toxins that make them more virulent. Examples are Grampositive bacteria, like Staphylococcus aureus and Enterococcus spp., which frequently display multidrug resistance [4] and Gram-negative Stenotrophomonas maltophilia. Infectious microbial disease remains a pressing problem worldwide, because microbes have resisted prophylaxis or therapy longer than any other form of life. In recent decades, problems of multidrug-resistant microorganisms have reached an alarming level in many countries around the world. A number of recent clinical reports describe the increasing occurrence of methicillin resistant Staphylococcus aureus (MRSA) which is most disturbing cause of nosocomial infections in developed countries [5,6]. In particular, increasing drug resistance among gram-positive bacteria such as Staphylococci, Enterococci and Streptococci is a significant health matter [7]. This has stimulated the scientists for the development of novel molecules to combat these illnesses. Moreover, methicillin resistant S.aureus (MRSA) represents a therapeutic problem of increasing importance, especially in hospital patients [8,9]. Benzimidazoles are remarkably effective compounds both with respect to their inhibitory activity and their favorable selectivity ratio. Extensive biochemical and pharmacological studies have confirmed that benzimidazole molecules are effective against various strains of microorganisms. Azole class of drugs particularly fused imidazoles occupy prominent place in medicinal chemistry because of their broad spectrum of pharmacological activities such as anti-inflammatory, analgesic, anticancer, antiulcer, antimicrobial, antiviral, pesticidal, cytotoxicity and anti-arrhythmic [10-13] activities. Infections caused by these microorganisms pose a serious challenge to the medicinal community and the need for an effective therapy has led to search for novel anti-microbial agents. During the last 20 years quantitative structure activity relationship (QSAR) models have gained an extensive recognition in physical, organic, analytical, pharmaceutical and medicinal chemistry. The success of the QSAR approach can be explained by the insight offered based on the structural determination of chemical properties, and the possibility to estimate the properties of new chemical compounds without the need to synthesize and test them among the homologous series [14].
Data on the in vitro antimicrobial activity of substituted benzimidazole derivatives having potent activity against MRSA of various derivatives belonging to this class are available [15] in the literature, enabling us to attempt an investigation on the quantitative structure activity relationships (QSAR) that govern their biological activities. We exploit this chemical moiety further to build a QSAR model to analyze the structure-activity relationship with the help of molecular descriptors. Such a model will provide insights into the kind of chemical groups that must be added or removed at a particular position of the common moiety to develop it into a potent therapeutic.
All computations and molecular modeling studies were carried out on a Windows XP workstation using the molecular modeling software package VLife Molecular Design Suite (V-Life MDS) version 3.5 [16].
Dataset and biological activity
A series of thirty one compounds substituted benzimidazole derivatives having potent activity against MRSA has been described in the literature was considered to perform the QSAR study [15]. The biological activity values represented as minimum inhibitory concentrations (MIC) were first converted to –log MIC (pIC50) in micromolar (Table 1) and were used as the dependent variable for the QSAR model. Table 1 shows the structure of thirty one such compounds along with their biological activity values.
| S.No | R1 | R2 | R3 | R4 | S. aureus | E. coli | B. subtilis | C. albicans |
|---|---|---|---|---|---|---|---|---|
| 1 | Cl | H | H |  |
4.3010 | - | 4.3010 | 4.3010 |
| 2* | Cl | Cl | H |  |
5.2041 | 4.3010 | 4.9030 | 4.9030 |
| 3 | Cl | Cl | Cl |  |
5.5058 | 4.3010 | 4.6020 | 4.3010 |
| 4* | Cl | Cl | NHCH(CH3)2 |  |
4.3010 | 4.3010 | 4.3010 | 4.3010 |
| 5 | Cl | Cl | Br |  |
5.5058 | 4.3010 | 4.6020 | 4.6020 |
| 6 | Cl | Cl | NH2 |  |
5.5058 | 4.3010 | 5.2041 | 5.2041 |
| 7* | Cl | H | NH2 |  |
4.6020 | 4.3010 | 4.6020 | 4.6020 |
| 8 | Cl | OC2H5 | 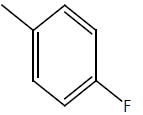 |
 |
5.2041 | 4.3010 | 4.6020 | 4.3010 |
| 9 | Cl | OC2H5 | 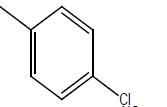 |
 |
4.3010 | 4.3010 | 4.6020 | 5.2041 |
| 10* | Cl | OC2H5 | 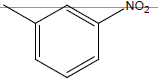 |
 |
4.3010 | 4.3010 | 4.6020 | 5.2041 |
| 11 | Cl | OC2H5 | 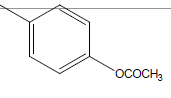 |
 |
4.3010 | 4.3010 | 4.3010 | 5.2041 |
| 12 | Cl | Cl | 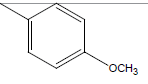 |
 |
4.6020 | 4.3010 | 4.6020 | 4.9030 |
| 13 | Cl | Cl | 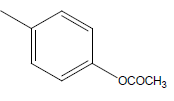 |
 |
4.3010 | 4.6020 | 5.2041 | 5.2041 |
| 14* | Cl | Cl | 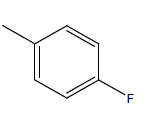 |
H | 5.5058 | 4.3010 | 4.3010 | 4.9030 |
| 15 | Cl | Cl | 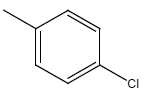 |
H | 5.5058 | 4.3010 | 5.2041 | 4.9030 |
| 16 | Cl | Cl | 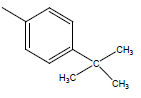 |
H | 5.5058 | 4.3010 | 4.6020 | 4.6020 |
| 17 | Cl | Cl | 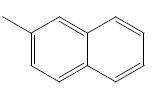 |
H | 4.3010 | 4.3010 | 4.6020 | 5.2041 |
| 18 | Cl | Cl | 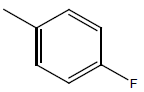 |
 |
4.6020 | 4.3010 | 4.3010 | 4.6020 |
| 19 | Cl | Cl | 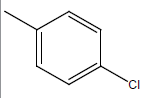 |
 |
4.3010 | 4.6020 | 4.6020 | 4.9030 |
| 20 | Cl | Cl | 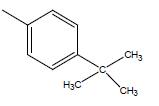 |
 |
5.2041 | 4.3010 | 4.6020 | 4.6020 |
| 21 | Cl | Cl | 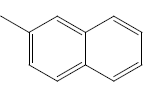 |
 |
4.3010 | 4.3010 | 4.6020 | 4.9030 |
| 22 | Cl | H | 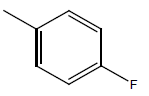 |
 |
4.9030 | 4.3010 | 4.9030 | 4.9030 |
| 23* | Cl | H | 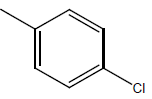 |
 |
4.3010 | 4.6020 | 4.6020 | 4.6020 |
| 24 | Cl | H | 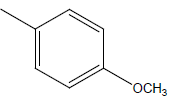 |
 |
4.3010 | 4.6020 | 4.6020 | 4.9030 |
| 25 | Cl | H | 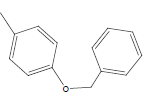 |
 |
5.5058 | 4.6020 | 4.3010 | 4.6020 |
| 26 | Cl | H | 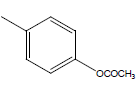 |
 |
4.3010 | 4.3010 | 4.3010 | 4.3010 |
| 27 | Cl | H | 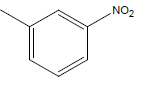 |
 |
4.3010 | 4.3010 | 4.6020 | 4.9030 |
| 28 | Cl | H | 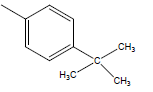 |
 |
4.3010 | 4.3010 | 4.3010 | 4.9030 |
| 29 | Cl | H | 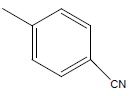 |
 |
4.3010 | 4.6020 | 4.3010 | 5.2041 |
| 30 | Cl | H | 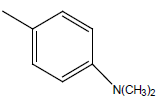 |
 |
4.9030 | 4.3010 | 4.6020 | 4.9030 |
| 31 | Cl | H | 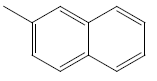 |
 |
4.6020 | 4.3010 | 4.6020 | 5.2041 |
*test set
Table 1: Structure, and biological activity of benzimidazole derivatives as Antibacterial and antifungal activities.
Optimization and selection of training and test set
Compounds were sketched using the 2D draw application and converted to 3D structures. Energy minimization and geometry optimization were conducted using Merck molecular force field [17] and atomic charges, maximum number of cycles were 1000, convergence criteria (RMS gradient) was 0.01 and medium’s dielectric constant of 1 by batch energy minimization method. The sphere exclusion method [18] was adopted for division of training and test data set comprising of 25 and 6 molecules, respectively.
Calculation of 2D descriptors
2D-QSAR study requires the calculation of molecular descriptors; almost 239 physicochemical descriptors were calculated by QSAR Plus module within VLife MDS. The invariable descriptors (descriptors that are constant for all the molecules) were removed, as they do not contribute to the QSAR. 2D descriptors were calculated which encoded different aspects of molecular structure and consists of electronic, thermodynamic, spatial and structural descriptors, e.g., retention index (chi),atomic valence connectivity index (chiV), path count, chain path count, cluster, path cluster, element count, estate number, semi-empirical, molecular weight, molecular refractivity, slogP, and topological index.
Model validation
Models generated by 2D -QSAR studies were cross validated using standard LOO procedure [19]. The cross validated r2 (q2) value was calculated using equation-1,where yi and yi are the actual and predicted activities of the ith molecule, respectively, and ymean is the average activity of all molecules in the training set. The q2 was calculated using Eq. (1).
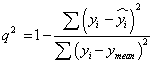 (1)
(1)
The predicted r2 (pred_r2) value was calculated using equation-2, where yi and yi are the actual and predicted activities of the ith molecule in test set, respectively, and ymean is the average activity of all molecules in the training set. The pred_r2 value is calculated as follows (Eq. (2))
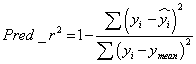 (2)
(2)
Some statistically significant QSAR models were chosen for discussion. The predicted (LOO) activities of the compounds by the above models are shown in Table 2.
| Parameter | T_N_N_1 | SssNHE-index | SsssCHcount | SsOHE-index | SssOE-index |
|---|---|---|---|---|---|
| T_N_N_1 | 1.0000 | ||||
| SssNHE-index | 0.3298 | 1.0000 | |||
| SsssCHcount | 0.2476 | 0.4731 | 1.0000 | ||
| SsOHE-index | 0.4764 | 0.5474 | 0.6943 | 1.0000 | |
| SssOE-index | 0.3794 | 0.4794 | 0.6891 | 0.8425 | 1.0000 |
Table 2: Correlation matrix between descriptors present in the best QSAR model -1.
pIC50 (S.aureus)=0.7842(± 0.1635) T_N_N_1 + 0.5882(±0.0473) SssNHE-index+0.3384 (±0.0685) SsssCHcount +0.5965(±0.0491) SsOHE-index +0.6083(±0.1273) SssOE-index
Ntraining = 25, Ntest= 6, r2= 0.8801, q2= 0.7269, F test =36.854, pred_r2 = 0.7843
The statistically significant penta-parametric model with principle component regression method with coefficient of determination (r2) = 0.8801 is capable of explaining 88% of variance in the observed activity values. The activity contribution chart for 2D QSAR model is shown in Figure 1a and plots of observed vs. predicted values of pIC50 are shown in Figure 1b.
pIC50 (E.coli) =-0.6518(±0.0375) SsFE-index +0.3885 (± 0.0331) SssCH2E-index
Ntraining = 25, Ntest= 5, r2= 0.8150, q2= 0.7135, F test=26.542, pred_r2 =0.7003
Model–2 shows good squared correlation coefficient (r2) of 0.8150 explains 81% variance in biological activity. Another parameter for predictivity of test set compound is Pred_r2 =0.7003, which is showing good external predictive power of the model.
pIC50 (B.subtilis) =0.4398(± 0.0437) T_2_C_1+0.8821 (± 0.1437) SdsCHcount
Ntraining =25, Ntest= 6, r2= 0.7565, q2= 0.7117, F test =25.654, pred_r2 = 0.6788
Model –3 shows good squared correlation coefficient (r2) of 0.7565 explains 75% variance in biological activity. Cross validated squared correlation coefficient of this model was 0.7117, which shows the good internal prediction power of this model. In this QSAR model 3 the positive coefficient of T_2_C_1, and SdsCHcount showed that increase in the values of these descriptors are beneficial for the activity.
pIC50 (C.albicans) =0.3873(± 0.0583) SddsN (nitro) count-0.1457(± 0.0231) SdsNE-index
Ntraining=25, Ntest= 6, r2= 0.7143, q2 = 0.6538, F test =21.864, pred_r2 = 0.6898
Model-4 explains 71% variance in biological activity. It also has an internal (q2) and external (pred_r2) predictive ability of ~65% and ~69% respectively.
Model-1 showed that the descriptor SsOHE-index which is topological state indices for number of –OH group connected with one single bond suggested at R1 substitution site, branching with hydroxyl atom is detrimental for activity. T_N_N_1 (count of number of Nitrogen atoms (single double or triple bonded) separated from any other nitrogen atom (single double or triple bonded) by one bonds in a molecule) reveals that (-NH) groups should be directly attached with benzimidazole ring at R3 position. The positive contributing SssNHEindex indices for the number of –NH groups connected with two single bonds. Its contribution in the model implies that it will decreases in potency R3 position. SsssCHcount indicates the presence of aliphatic ring substituents at the R2 and R3 position of the benzimidazole derivatives which is detrimental for the activity. SssOE-index indices for number of oxygen atom connected with two single bonds showed positive effect indicated that the antibacterial activity was increased with the presence of methoxy groups. The correlation matrix between the physico-chemical parameters and the biological activity for the models 1 is presented in Table 3. Model-2 reveals that SsFE-index is suggests that presence of fluorine atoms in the molecule as indicated by the good activity of molecules having fluorine atoms benzimidazole nucleus at R2 and R3 will lead to improved activity. The descriptor SssCH2E-index, estate indices for the number of CH2 groups attached with two single bonds showed that phenyl ring positively influences benzimidazole derivatives. Model-3 reveals that the descriptor SdsCHcount (total number of –CH group connected with one double and one single bond) indicates the relationship with reference to variation at R2 and R3 position as in case of compounds 1, 2, 7, and 22-31 benzimidazole ring and suggests that increase in length of – CH atoms chain on that substitution site is favorable for the activity. The descriptor T_2_C_1 indicates that presence of substituent’s with direct attachment of carbon on aromatic ring favorable for the activity. Model-4 indicate SddsN (nitro) count suggests the total number of nitro group connected with one single and double bonds R3 position will lead to improved activity. Descriptor SdsNE-index (~27%) indices for number of nitrogen atom connected at two double bond and one single bond is directly proportional to the activity. It shows that Nitro group in benzimidazole derivatives is essential for the activity at R3 site.
| Com. | 2D model (S.aureus) | 2D model (E.coli) | 2D model (B.subtilis) |
2D model (C.albicans) |
||||
|---|---|---|---|---|---|---|---|---|
| Pred. | Res. | Pred. | Res. | Pred. | Res. | Pred. | Res. | |
| 1 | 4.1614 | 0.139 | - | - | 4.4149 | -0.113 | 4.1710 | 0.13 |
| 2 | 5.0415 | 0.163 | 4.1181 | 0.182 | 4.8195 | 0.083 | 4.7424 | 0.160 |
| 3 | 5.2758 | 0.230 | 4.3845 | -0.083 | 4.4664 | 0.135 | 4.1916 | 0.1094 |
| 4 | 4.2396 | 0.061 | 4.2677 | 0.033 | 4.3655 | -0.064 | 4.2724 | 0.028 |
| 5 | 5.5113 | -0.005 | 4.1059 | 0.195 | 4.5762 | 0.025 | 4.4922 | 0.109 |
| 6 | 5.5238 | -0.018 | 4.2015 | 0.099 | 5.2134 | -0.009 | 5.0338 | 0.170 |
| 7 | 4.7949 | -0.192 | 4.0529 | 0.248 | 4.4928 | 0.109 | 4.5810 | 0.021 |
| 8 | 5.0976 | 0.106 | 4.3951 | -0.094 | 4.5058 | 0.096 | 4.2968 | 0.004 |
| 9 | 4.2312 | 0.069 | 4.4792 | -0.178 | 4.5374 | 0.064 | 5.1758 | 0.028 |
| 10 | 4.1771 | 0.123 | 4.1755 | 0.125 | 4.4997 | 0.102 | 5.1970 | 0.007 |
| 11 | 4.2668 | 0.034 | 4.2264 | 0.074 | 4.4852 | -0.184 | 5.1438 | 0.060 |
| 12 | 4.7033 | -0.10 | 4.7117 | -0.109 | 4.6923 | -0.090 | 4.9707 | -0.067 |
| 13 | 4.2194 | 0.08 | 4.1613 | 0.139 | 5.2145 | -0.010 | 5.2505 | -0.046 |
| 14 | 5.3658 | 0.14 | 5.6618 | -0.156 | 4.1617 | 0.139 | 4.8853 | 0.017 |
| 15 | 5.2602 | 0.2456 | 5.2723 | 0.233 | 5.2247 | -0.020 | 4.9101 | -0.007 |
| 16 | 5.4896 | 0.0162 | 5.6513 | -0.145 | 4.5481 | 0.053 | 4.5683 | 0.033 |
| 17 | 4.1642 | 0.1368 | 4.1754 | 0.125 | 4.5270 | 0.075 | 5.2839 | -0.0798 |
| 18 | 4.4783 | 0.1237 | 4.1818 | 0.119 | 4.2314 | 0.069 | 4.6913 | -0.089 |
| 19 | 4.1982 | 0.1028 | 4.4491 | 0.152 | 4.7846 | -0.182 | 4.8214 | 0.081 |
| 20 | 5.1195 | 0.0846 | 4.1131 | 0.187 | 4.4772 | 0.124 | 4.6709 | -0.068 |
| 21 | 4.3675 | -0.066 | 4.5685 | -0.267 | 4.6248 | -0.022 | 4.8349 | 0.068 |
| 22 | 4.7382 | 0.164 | 4.0483 | 0.252 | 5.1087 | -0.205 | 4.9347 | -0.031 |
| 23 | 4.149 | 0.152 | 4.7768 | -0.174 | 4.3866 | 0.215 | 4.6431 | -0.041 |
| 24 | 4.1984 | 0.102 | 4.5659 | 0.036 | 4.7867 | -0.184 | 4.8528 | 0.050 |
| 25 | 5.4147 | 0.091 | 4.5541 | 0.047 | 4.6966 | -0.395 | 4.5598 | 0.042 |
| 26 | 4.0674 | 0.233 | 4.1014 | 0.199 | 4.0414 | 0.259 | 4.4647 | -0.163 |
| 27 | 4.2531 | 0.047 | 4.1519 | 0.149 | 4.5728 | 0.029 | 4.8908 | 0.012 |
| 28 | 4.5357 | -0.234 | 4.2542 | 0.046 | 4.1919 | 0.109 | 4.8257 | 0.077 |
| 29 | 4.5129 | -0.211 | 4.4796 | 0.122 | 4.1773 | 0.12 | 5.2294 | -0.025 |
| 30 | 4.6868 | 0.216 | 4.1253 | 0.175 | 4.7136 | -0.111 | 4.9106 | -0.007 |
| 31 | 4.4994 | 0.102 | 4.1596 | 0.141 | 4.3899 | 0.212 | 5.1616 | 0.042 |
Table 3: Observed and Predicted Activities (pIC50) for the Training and Test Set Compounds.
QSAR studies were performed to design new more potent benzimidazole compounds for antimicrobial activity. From the detailed analysis of the results of above studies, we conclude that antibacterial activity of the compounds significantly depends on the electronic effect of hydroxyl and fluoro groups. QSAR model indicated that the antibacterial activity was increased with the presence of methoxy groups and fluorine atoms benzimidazole nucleus at R1, R2 and R3 will lead to improved activity. Variables in the equation revealed that thermodynamic, electronic, structural and molecular shape analysis descriptors contribute significantly to the antimicrobial activity. These trends should prove to be an essential guide for the future work. Thus, the results obtained from QSAR study gives a hypothetical image to design new potent antimicrobial activity.
The author wishes to express gratitude to V-life Science Technologies Pvt. Ltd for providing the trail version software for the study.