
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2024)Volume 7, Issue 2
Objective: The potential mechanisms of NF1, Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and Neurofibromatosis 1 (NF1) expressions in Glioblastoma Multiforme (GBM) have been poorly elucidated. Risk-score analysis combined with clinic pathological characteristics to assess the prognostic value and immunotherapy efficacy of 6-genes signature.
Methods: We performed the whole exosome sequencing profiling on samples form patients with GBM. The association of PIK3CA and NF1 expression within the GBM have received cross-validations by Mendelian Randomization (MR) methods and the risk-score analysis of 6-gene signature.
Results: MR study demonstrated that PIK3CA and NF1 expressions were closely related with the risk of GBM patients by the Inverse Variance Weighting (IVW) method and then the construction of 6-gene signature was classified into high-risk and low-risk groups through the median risk-score of regression formula to predict prognosis of GBM patients. Kaplan-Meier survival showed the overall survival of distinct high and low-risk groups. Receiver Operating Characteristic (ROC) curve analysis indicated the value of predictive performance of two different risk groups. In line with our Whole Exome Sequencing reporters (WES), our findings showed that the mutation of PIK3CA and NF1 was significantly associated with prognostic signature of GBM.
Conclusion: Taken together, our MR analysis indicated the correlation between PIK3CA and NF1 expression and GBM disease, which provided a key basis for the precise prevention of the genetic mutation in the occurrence and development of GBM. The established PIK3CA and NF1 alteration-related prognostic signature was involved well in prognosis prediction as well as closely linked with immunotherapy responses.
Mendelian randomization; Glioblastoma multiforme; PIK3CA; NF1; Ankylosing spondylitis
CR: Complete Response; PR: Partial Response; SD: Stable Disease; PD: Progressive Disease
In human brain, Glioblastoma Multiforme (GBM) is the one of most malignant brain cancer within the Central Nervous System (CNS) [1-3]. The gross total surgical resection and standard postoperative chemo radiotherapy is our current effective treatment to improve survival of patients with GBM. However, GBM with high therapeutic resistance and poor prognosis, is the most common intracranial primary malignant human tumour [4-6]. At present, our therapeutic methods have limited effect on improvement of the prognosis of GBM cases. It has reported that Food and Drug Administration (FDA) has approved two drugs to treat GBM patients including Temozolomide (TMZ) and bevacizumab, but the prognostic results indicated that only 15 months of median survival and less than 5% of five-year survival rate [7-11].
The stromal cells, immune cells and the Extracellular Matrix (ECM) and other secreted molecules consisted on the Tumour Microenvironment (TME) which was significantly associated with the molecular phenotypic heterogeneity in the GBM and then took effects on the regulation of the mechanism of molecular phenotypic plasticity of GBM. Overall, the TME took a vital role in the improvement survival in the malignant progression of GBM. Moreover, GBM cells secreted diverse cytokines or chemokines or expressed immunosuppressive molecules Programmed Cell Death Ligand 1 (PD-L1) to exert immunosuppressive effects [12-14].
Because the routine treatment strategy for GBM patients is not fully, some new methods, such as immune therapy are applied in clinical practice. The development of immunotherapy for GBM functioned as a novel treatment to manipulate the immune system to attack tumor cells with lower adverse effects. Overall, understanding the heterogeneity and diverse functions of brain cancer is necessary to design immunotherapeutic strategies for GBM.
To gain insights into finding more efficient treatment approaches against GBM. We performed the whole exome sequencing profiling on samples form patients with GBM. Moreover, as we known that mendelian randomization mainly was performed as a genetic epidemiological approach to explore the potential causality of exposure and outcome through genetic variants including Single-Nucleotide Polymorphisms (SNPs). Due to traditional observational epidemiological method are more likely to confounders and reverse causation bias, the MR analysis function as a genotype of an individual is certain and unchanged to improve confounders and reverse causation bias [15].
There are no MR analysis inferring the potential relationship of gene expression in glioblastoma multiform to data. Thus, the twosample MR approach to infer whether the genetically predicted PIK3CA and NF1 gene expression were linked with the risk of the glioblastoma multiform. Furthermore, six Differentially Expressed Genes (DEGs) from mutant GBM were screened out and selected to construct a novel risk score model for prognosis and risk stratification in GBM cases based on The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) dataset. More in depth evaluation and validation for nomograms and ROC curves independently predicted the association between 6 genes signature and clinical traits and immunotherapy efficacy form GBM patient’s prognosis with moderate accuracy. Our studies demonstrated that targeting PIK3CA and NF1 could enhance ICB immunotherapy for glioma. Reinforcing PIK3CA and NF1 targeted therapy as a promising strategy.
Mendelian randomization analysis and the selection of instrumental variable
The summary statistics of PIK3CA and NF1 expression and GBM disease were from published Genome Wide Association Studies (GWAS) in predominantly European individuals mainly including males and females. Instrumental Variable (IV) of SNPs at genome-wide significance level was chose for PIK3CA and NF1 expression and GBM disease as genetic variants. We failed to found several SNPs with Linkage Disequilibrium (LD) in the sensitivity analysis. F-statistics was calculated and quantified the strength of the instrumental variable in order to evaluate IVs bias and the assumption of F>10 for MR study. In addition, the minimum effect size (80%) statistical power at a level of 5%.
Data acquisition and establishment of prognostic signature
Overall survival-associated several genes with prognostic power were performed to construct risk models and then divided into high-risk and low-risk groups based on TCGA-GBM cohort and GEO dataset. The prognostic survival and ROC curve analysis were performed to evaluate the performance of 6-gene signature in prognostic prediction. Then, the differentiation analysis of clinical characteristics including age and gender in high-risk and low-risk groups. We acquired the somatic mutation data and somatic copy number alterations of GBM patients based on The Cancer Genome Atlas Program (TCGA) dataset.
Gene set enrichment analysis functional enrichment analysis
We explore the differentiation of biological function within the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in high and low risk groups based on cluster profiler Gene Set Enrichment Analysis (GSEA) R package and then enrichment score was calculated with adjusted P-value less than 0.05 as statistically significant and cut-off of P<0.01.
The analysis of immunotherapy response to programmed cell death ligand-1
We conducted and testified the treatment response to programmed death ligand 1 also known as CD274 blockade through IMvigor210 cohort. A group of GBM patients was treated by immunotherapy with anti-PD-L1 therapy and the treatment response criteria.
The analysis of genetic alteration of PIK3CA and NF1
In order to know genetic alteration characteristics of PIK3CA and NF1, we logged into the cBioPortal website (https://www. cbioportal.org/). We mainly explored the module of the cancer types summary to acquire the Copy Number Alteration (CNA), gene mutated types and alteration frequency of PIK3CA and NF1. The protein structure like Three-Dimensional (3D) could be shown in the schematic diagram. And the genetic mutated sites of two genes also were found through the mutated module. Additionally, the overall survival, Disease Free Survival (DFS) and Disease Specific Survival (DSS) were compared from with genetic alteration to without genetic alteration via the comparison module and the results were displayed by Kaplan-Meier plots with log-rank P-value.
Statistical analysis
Because of our data form published studies and public dataset, no additional ethical approval from an institutional review board was required.
The two-sample Mendelian Randomization (MR) study was performed to evaluate the potential causal association of PIK3CA and NF1 expression with GBM disease through mendelian randomization and Mendelian Randomization Pleiotropy Residual Sum and Outlier (MRPRESSO) R packages of version 3.6.
Inverse Variance Weighted (IVW) approach was conducted for the consistent assessment of the correlation between exposure and the risk of outcomes as IVs were not pleiotropic [15]. We estimated the heterogeneity through individual SNPs based on Cocchran’s Q statistics. When there existed no substantial heterogeneity (p<0.05), we used the fixed-effects model or we conducted the random-effects model [16].
The sensitivity analysis was used to prove the robustness and accuracy of our study. We also used the weighted median approach for a consistent assessment of the overall effect as the IVs were valid (more than 50%), which performed better in reducing the bias of the causal effect assessment than the IVW approach [17]. Then, we conducted the MR-egger regression to evaluate whether the instrumental SNPs were multi-effect. Which was performed to measure the directional pleiotropy and attain the intercept [18]. Additionally, statistical significance was set at a P-value, which was corrected for two exposures and one outcome through Bonferroni approach. Probability value (P-value) low than 0.05 or more than Bonferroni-corrected threshold, which was regarded suggestive evidence for an underlying and potential causal correlation. All statistical analyses of our data were twosided analysis and the statistical analyses in our manuscript were performed in R software.
MR analysis of NF1 and PIK3CA gene expression
An overview of our manuscript design is given in Figure 1. MR analyses indicated the association between NF1 and PIK3CA with GBM (Figure 2). We used the fixed effect IVW model for MR analysis through Cochran’s Q test. A higher PIK3CA expression genetically predicted was closely related with a lowrisk of GBM by the IVW method (OR: 0.176, 95% CI: 0.056-0.556, P=0.003). The sensitivity analysis gave similar outcomes (OR: 0.279, 95% Class Interval (CI): 0.074-1.047, P=0.059) by weighted median; OR: 0.316, 95% CI: 0.043-2.320, P=0.321 by simple mode; OR: 0.312, 95% CI: 0.059-1.650, P=0.242 by weighted mode. There was no significant evidence for the association between NF1 expression and GBM by using IVW method (OR: 0.871, 95% CI: 0.486-1.562, P=0.643) (Figure 2). In addition, we observed high expression of NF1 in Ankylosing Spondylitis (AS) by the IVW method (OR: 0.999, 95% CI: 0.999-1.000, P=0.042), which indicated genetic causal relationship with AS. Moreover, there existed similar tendency by weighted median, simple mode or weighted mode methods. MR Egger-regression showed no evidence of potential directional pleiotropy between PIK3CA (P for intercept=0.972) but NF1 (P for intercept=0.379). Here we observed whether the SNPs are horizontally pleiotropic based on MR-Egger method because of MR-Egger value required more than three SNPs. The leave-one-out analysis demonstrated whether the causal estimate was driven by any single SNP, which indicated a consistent inverse correlation between PIK3CA and NF1 expression and risk of GBM, as well as statistically significant correlation between NF1 expression with AS. Overall estimates were shown no disproportionately affected by any individual SNP. The effect estimates of MR analyses for the inverse correlation of NF1 and PIK3CA with GBM diseases. The results showed the positive association between NF1 level with AS disease. Scatter plot and funnel plot of NF1 and PIK3CA expression in GBM were shown no evidence of horizontal pleiotropy in, as well as no statistically significant correlation between NF1 expression with AS.
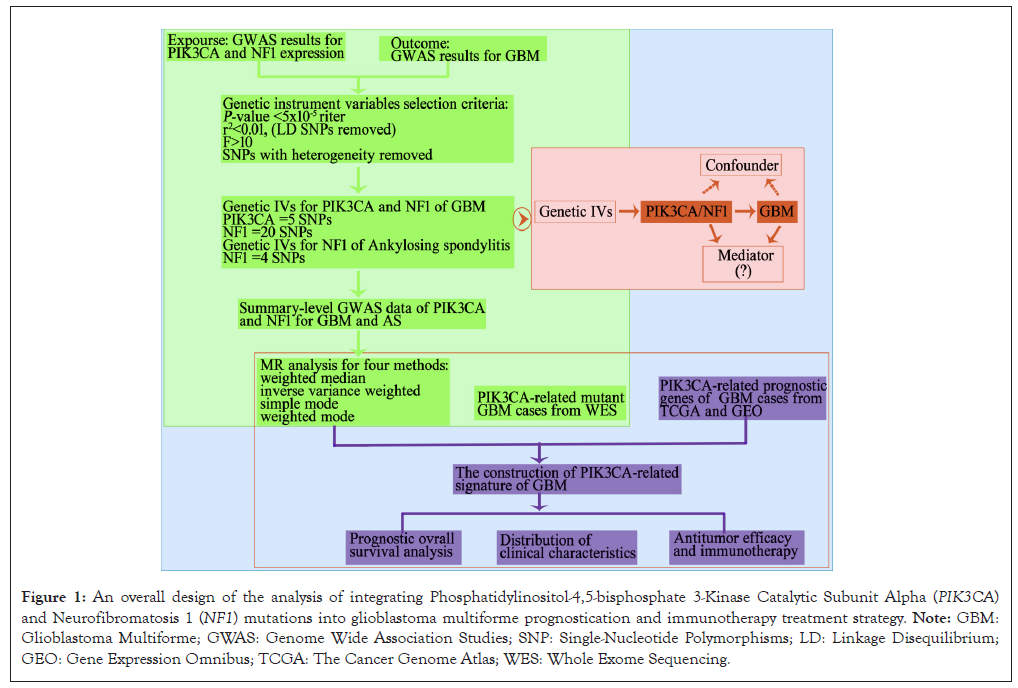
Figure 1: An overall design of the analysis of integrating Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and Neurofibromatosis 1 (NF1) mutations into glioblastoma multiforme prognostication and immunotherapy treatment strategy. Note: GBM: Glioblastoma Multiforme; GWAS: Genome Wide Association Studies; SNP: Single-Nucleotide Polymorphisms; LD: Linkage Disequilibrium; GEO: Gene Expression Omnibus; TCGA: The Cancer Genome Atlas; WES: Whole Exome Sequencing.
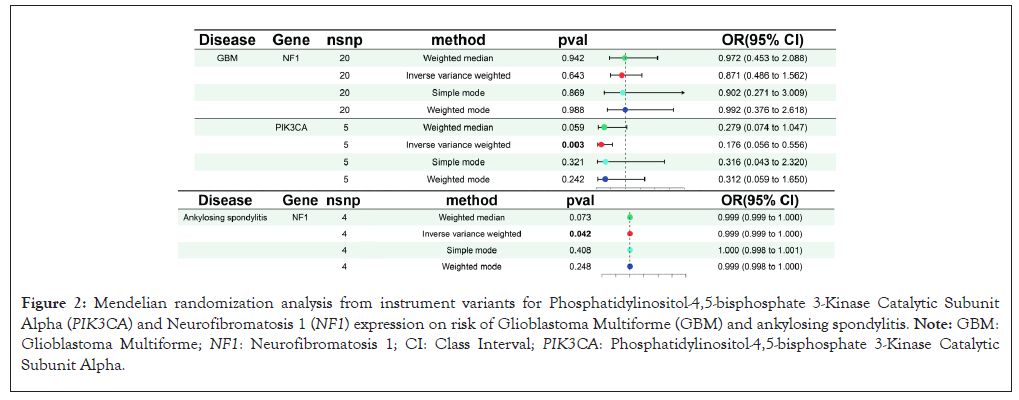
Figure 2: Mendelian randomization analysis from instrument variants for Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and Neurofibromatosis 1 (NF1) expression on risk of Glioblastoma Multiforme (GBM) and ankylosing spondylitis. Note: GBM: Glioblastoma Multiforme; NF1: Neurofibromatosis 1; CI: Class Interval; PIK3CA: Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha.
The construction of 6-genes signature and clinical prognostic model
Six genes with were closely related with overall survival within the GBM cases through difference multiple and significance threshold from TCGA and GEO dataset. Subsequently, we first constructed a 6-gene signature through univariate prognostic analysis. The 6-gene signature was divided into high-risk and low-risk groups (Figures 3a and 3b). The time-dependent ROC curve analysis demonstrated that the area under the Area Under Curve (AUC) values for 1, 3 and 5-years overall survival were 0.622, 0.743 and 0.798 respectively (Figure 3c). Importantly, 6-gene signature displayed the strongest predictive power for overall survival in GBM was 0.798 and other age curve, gender or grade were 0.668, 0.321 and 0.5 respectively (Figure 3d). We also observed that patient’s age and gender were closely associated with diverse risk groups. Such as, female GBM patients who more than 65 existed in high-risk group compared to others (Figure 4a). The Glioblastoma Multiforme (GBM) patients were stratified into high-risk and low-risk groups based on the mendelian cut-off value of risk-score. The analysis of 6-gene signature prognostic survival and clinical characteristics including age and gender of GBM patients in high- and low-risk groups.
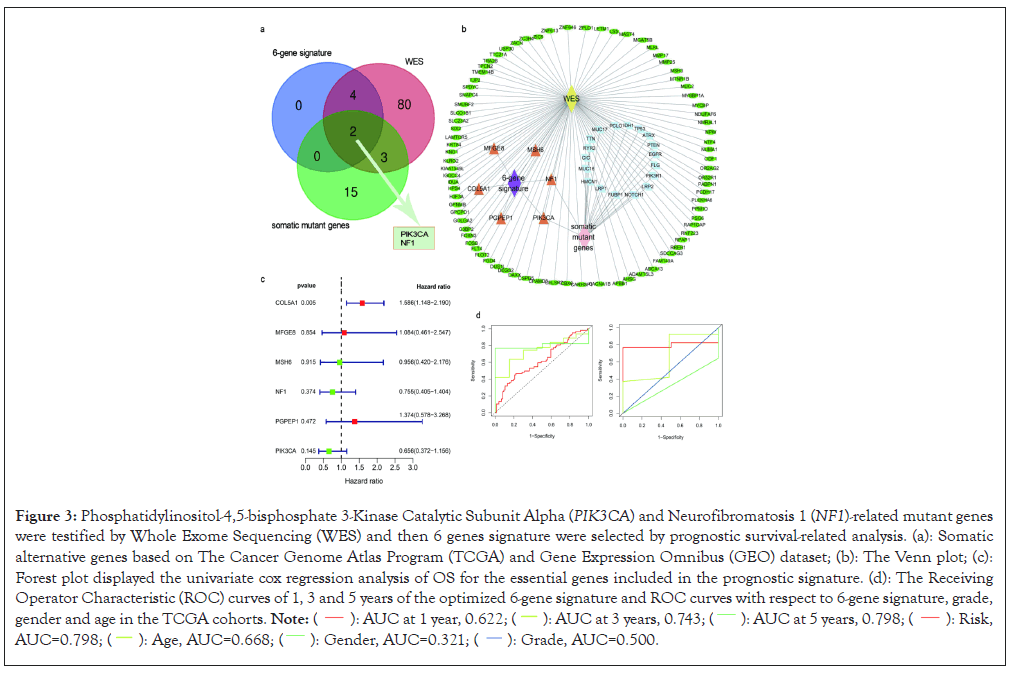
Figure 3: Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and Neurofibromatosis 1 (NF1)-related mutant genes
were testified by Whole Exome Sequencing (WES) and then 6 genes signature were selected by prognostic survival-related analysis. (a): Somatic
alternative genes based on The Cancer Genome Atlas Program (TCGA) and Gene Expression Omnibus (GEO) dataset; (b): The Venn plot; (c):
Forest plot displayed the univariate cox regression analysis of OS for the essential genes included in the prognostic signature. (d): The Receiving
Operator Characteristic (ROC) curves of 1, 3 and 5 years of the optimized 6-gene signature and ROC curves with respect to 6-gene signature, grade,
gender and age in the TCGA cohorts. 

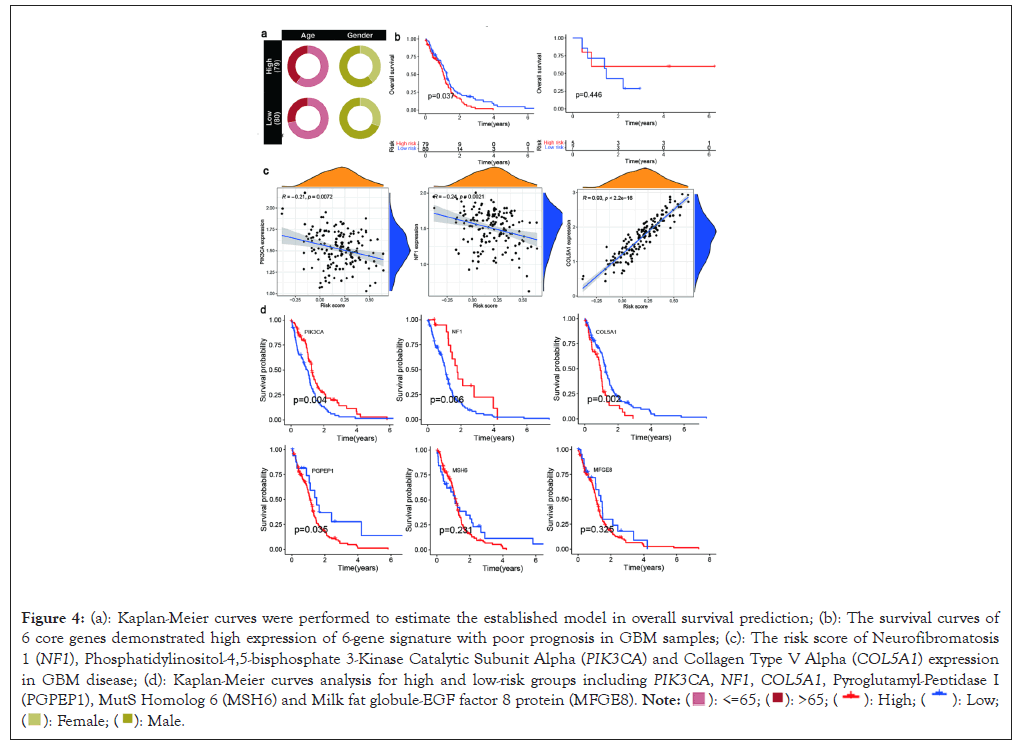
Figure 4: (a): Kaplan-Meier curves were performed to estimate the established model in overall survival prediction; (b): The survival curves of
6 core genes demonstrated high expression of 6-gene signature with poor prognosis in GBM samples; (c): The risk score of Neurofibromatosis
1 (NF1), Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and Collagen Type V Alpha (COL5A1) expression
in GBM disease; (d): Kaplan-Meier curves analysis for high and low-risk groups including PIK3CA, NF1, COL5A1, Pyroglutamyl-Peptidase I
(PGPEP1), MutS Homolog 6 (MSH6) and Milk fat globule-EGF factor 8 protein (MFGE8). 
The ROC curve indicated that the risk score of the 6-gene signature made an excellent predictive accuracy. Furthermore, the prognostic survival of high and low-risk groups of GBM cases were significantly different based on TCGA dataset (P=0.037). The study demonstrated high-risk group with a poor overall survival compared with low-risk cases. But we failed to find the similar result through GEO dataset (Figure 4b). Interestingly, the gene expression of both NF1 (R=0.24, P<0.0021 and PIK3CA R=0.21, P<0.0072) were closely negative with high-risk group. In contrast, as shown in Figure 4c, the survival analysis of 6-genes signature demonstrated that the Collagen Type V Alpha (COL5A1) expression was significantly positive with high-risk group (R=0.93, P<2.2e-16). The six prognostic genes demonstrated that NF1 (P<0.004) and PIK3CA (P<0.006) expression were good at survival prognosis of GBM patients. Differently, other four genes were significantly associated with poor prognosis of GBM patients. Such as, the high expression of COL5A1 with lower survival rate (P<0.002) and high Pyroglutamyl-Peptidase I (PGPEP I) expression with worse survival (P<0.035). But we failed to find the expression of MutS Homolog 6 (MSH6) and Milk Fat Globule-EGF Factor 8 protein (MFGE8) (Figure 4d).
The analysis of genetic alteration of PIK3CA and NF1 genes
We explore the genetic alteration status of PIK3CA and NF1 expression in diverse tumor types of TCGA cohorts and the results showed the highest alteration frequency of two genes level appeared for patients with GBM and Low-Grade Gliomas (LGG) tumor types with ametelin as the primary type (Figure 5aⅠ). The “amplification” type of Copy Number Alteration (CNA) also existed the GBM and LGG tumor types. It was worth noting that the “deep deletion” type of CNA only expressed in the GBM and LGG tumor associated with the NF1 expression, which displayed an alteration frequency of less than 5% (Figure 5aⅡ). Moreover, the “multiple alterations” type of CNA as well as existed in GBM and LGG tumors linked with NF1 expression. The results presented the types and sites and case number of PIK3CA and NF1 genetic alteration. The missense mutation (number: 1366) of PIK3CA was the primary type of mutation and E545K/A/G alteration in the PI3Ka domain (Figure 5bⅠ), which could induce a frame shift mutation of the PIK3CA expression, translation form Glutaurine (E) to Lysine (K) or Alanine (A) or Glycine (G) at the 545 site of PIK3CA protein and the subsequent PIK3CA protein inframe (number: 43) and protein truncation (number: 2). The E545 site of PIK3CA presented in the 3D protein structure (Figure 5bⅡ). Moreover, we displayed that missense mutation of NF1 gene (number:15) was the type of genetic alteration (Figure 5cⅠ) and K1661Gfs*36 alteration in the Ras GTPase-Activating Proteins (RasGAP) domain, which could induce a frame shift mutation of the NF1 gene, translation from Lysine (K) to Glycine (G) at the 1661 site of NF1 protein and subsequent NF1 protein splice (number: 96), fusion (number: 6) and truncating (number: 350), but we cannot found the K1661 site in the 3D structure of NF1 protein (Figure 5cⅡ).
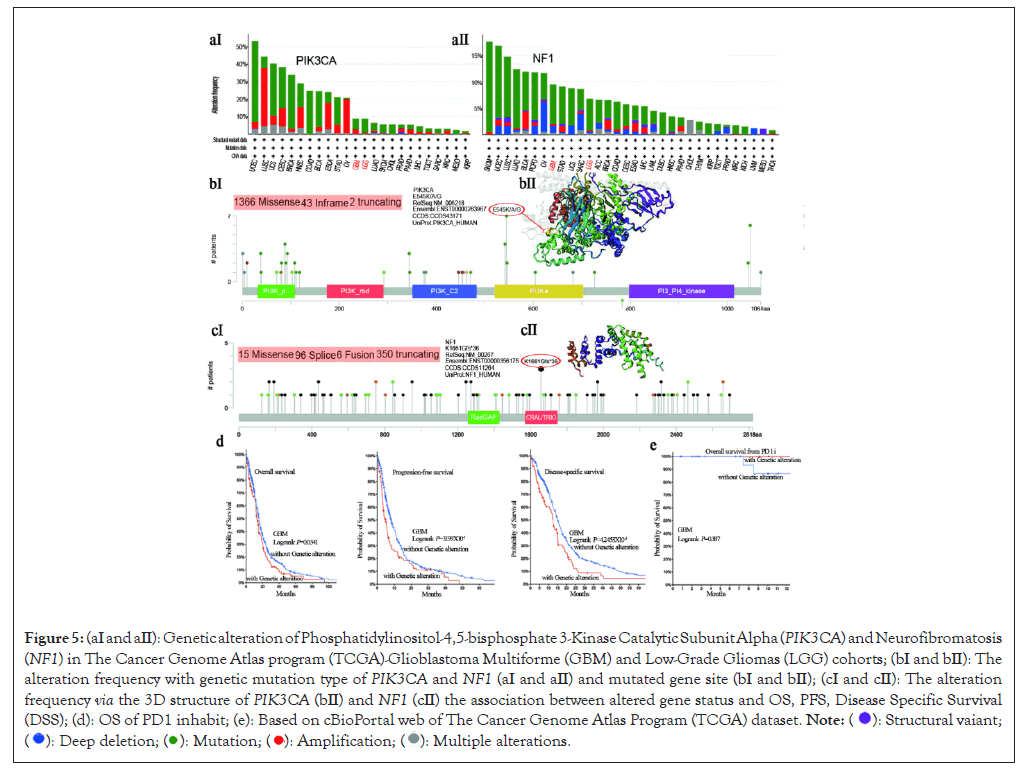
Figure 5: (aⅠ and aⅡ): Genetic alteration of Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) and
Neurofibromatosis (NF1) in The Cancer Genome Atlas program (TCGA)-Glioblastoma Multiforme (GBM) and Low-Grade Gliomas (LGG)
cohorts; (bⅠ and bⅡ): The alteration frequency with genetic mutation type of PIK3CA and NF1 (aⅠ and aⅡ) and mutated gene site (bⅠ and bⅡ); (cⅠ
and cⅡ): The alteration frequency via the 3D structure of PIK3CA (bⅡ) and NF1 (cⅡ) the association between altered gene status and OS, PFS,
Disease Specific Survival (DSS); (d): OS of PD1 inhabit; (e): Based on cBioPortal web of The Cancer Genome Atlas Program (TCGA) dataset. 
Importantly, we also investigated the potential relationship between genetic mutation and the clinical prognostic survival of samples with diverse cancers. Our results found that GBM cases with altered PIK3CA and NF1 was worse prognosis in overall (p=0.0341), DFS (p=0.003119) and DSS (p=0.0042453) (Figure 5d). In addition, we also explored the association between mutated PIK3CA and NF1 expression and PD1 inhabit treatment within the GBM tumor type and our data found that the OS with altered gene was better prognosis (p=0.387), which show the immunotherapy treatment may be good at GBM tumor (Figure 5e). More research needed to be represented.
The expression of cancer somatic genome of 6-genes signature and the prediction of immunotherapeutic benefit of 6-genes signature in the IMvigor210 cohort
High-risk and low-risk groups had the heterogeneity in the genetic mutation. The differences of somatic cells mutations demonstrated that two genes with mutation frequency in top-20 simultaneously appeared in the low-risk (left panel) and high-risk groups (right panel) respectively. The highest alteration frequency of PIK3CA (~11%) appeared for patients with GBM with “missense mutation” as the primary type in the low-risk group compared the high-risk group with (~9% frequency). In contrast, an alteration frequency of NF1 (~15% frequency) in the high-risk group more than in the low-risk group (~9% frequency).
Emerging literature have reported that genetic alteration could respond to immune-checkpoint therapy, which indicated that 6-genes signature might be more useful for predicting prognostic benefits in GBM patients with immunotherapy treatment [4,19,20]. The distributions of somatic alterations and different patterns within the terms of gene mutations found two of twenty variant mutated genes, which were closely linked to MR analysis in the GBM cases. Clinical and pre-clinical studies have found the correlations between individual mutated genes and response or resistance to Intracranial Pressure (ICP). Such as PIK3CA and NF1, which may exclusively associate with sensitivity or resistance in GBM samples.
Furthermore, we evaluated with ROC analysis in the IMvigor210 cohort and observed a predictive advantage of predictive value through pROC package (95% CI: 0.487-0.645). The prognostic value of high-risk and low-risk groups for immune checkpoint therapy were performed to explore the clinical survival (P=0.016) of GBM patients in the IMvigor210 cohorts. Our results demonstrated that GBM patients with low-risk group mad significantly longer prognostic survival than those with lower-risk group in the IMvigor210 cohort. In good agreement with predicted outcomes of anti-PD-L1 treatment, we obtained that predictive value of low-risk group were more likely to benefit from immune checkpoint therapy in to IMvig-or210 cohort. These findings might give a new perspective to explore the mechanism and reveal individual genetic mutations and their role in human tumor immunity and immunotherapy.
The analysis of GSEA functional annotation and cancer somatic genome of 6-genes signature
Gene Set Enrichment Analysis (GSEA) mainly demonstrated the enrichment of 6-genes signature was linked to brain-related pathway like Parkinson, neuroactive ligand receptor interaction, cell cycle, butanoate metabolism, oxidative phosphorylation in the low-risk group and chemokine signaling pathway and so on in the high-risk group. Above pathway results were consistent with our analysis, more in-depth research was performed for our analysis. In addition, we also investigated the protein expression characteristics of PIK3CA and NF1 genes by analyzing the Immunohistochemistry (IHC) images from the Human Platelet Antigen (HPA) dataset, which results showed the deeply stained proteins in GBM stroma. These data indicated that PIK3CA and NF1 might be biomarkers within the GBM.
Averaging one year following diagnosis, which accounted for 82% patients with malignant glioma (GBM-GBM 2). There remain lacked of effective immunotherapeutic successes for the treatment of GBM and some factors challenge the current clinical trials of immunotherapy in GBM, such as tumour heterogeneity, drug penetration into Blood-Brain Barrier (BBB) and immunosuppressive Tumor Microenvironment (TME). Thus, we utilized the Whole Exome Sequencing (WES) data in combination with bioinformatics approach to construct a signature model for prognosis prediction and risk stratification in GBM. The high and low-risk score model offered a potential and novel theoretical basis for choice of immunotherapy approach other than chemotherapy drugs like TMZ in GBM, which might provide a novel and effective way for extension of GBM patients survival time, particularly for no response to chemotherapy drugs.
Messenger Ribo Nucleic Acid (RNA) levels of Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Beta (PIK3CB) and Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) were significantly higher than those of Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Delta (PIK3CD) and Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic Subunit Gamma (PIK3CG) in GBM cell lines. PIK3CA (the p110 α subunit of PI3K) or Phosphoinositide-3-Kinase Regulatory Subunit 1 (PIK3R1) was amplified or mutated in approximately 15% of GBM and which could integrate the regulation of multiple GBM hallmarks by directly targeting the PIK3CA oncogene that is known to be important for GBM [21- 23].
GBM subtype was characterized by a high frequency of NF1 mutation or deletion, in combination with p53 mutations and loss of heterozygosity [24].
Recent sequencing results have demonstrated that NF1 carries point mutations in 14%-15% of GBMs, with an additional 9% showing deletion of the NF1 gene [25].
In the present study, the role of PIK3CA and NF1 expression played in the occurrence and prognosis of GBM by the integrative bioinformatics analysis and MR approach according to the report of Whole Exosomes (WES) from GBM patient. Firstly, a higher PIK3CA and NF1 expression per one SD was linked with a decreased risk of GBM by the IVW approach. Sensitivity analysis using alternative approaches demonstrated similar results. The MR-Egger regression showed the evidence of pleiotropy. Interestingly, we found that high NF1 expression per one SD was closely correlated with an increased AS. Our MR analysis provided convincing evidence for an inverse association between PIK3CA and NF1 expression and risk of GBM. More in depth research was warranted to elucidate the possible relationship between gene expression and risk. The assessment of clinical characteristics and prognostic survival by the 6-genes signature offered a predictor of survival in AS. The 6-genes signature was a biomarker for predicting prognostic survival in GBM and also designed to evaluate immunotherapy efficacy GBM patients based on TCGA and GEO cohort. 6-gene signature made of PIK3CA and NF1 genes, were divided GBM samples from TCGA and GEO cohort into high-risk and low-risk groups. The Kaplan-Meier curve indicated that GBM patients prolonged the survival and had better prognosis in the low-risk group compared with high-risk group. Importantly, 6-genes signature made a good performance in prognostic prediction and it also was involved in inference of clinic pathological characteristics. The construction of ROC curve of 6-genes signature functioned as a prognosis prediction. Subsequently, GSEA functional annotation revealed that multiple kinds of immune and brain related signalling pathways were enriched in the low-risk group. Consistent with a previous study about MR analysis of the association of PIK3CA and NF1 expression in GBM. The higher score group and stromal were correlated with worse overall survival after initial treatment in GBM. Thus, the analysis of novel and potential molecular targets in GBM is essential for the development of immune targeting therapies.
PIK3CA and NF1 made pivotal roles in regulating signal activation, which benefited the tumorigenesis, tumour development and progression. As the large-scale sequencing data indicated that the established PIK3CA hotspots could drive in vivo tumorigenesis and then the cohort of PIK3CA variants were screened and found in human GBM. PIK3CA is an important driver of malignant astrocytic brain tumour. It reported that the mutated or deleted of PIK3CA gene in about 40% of GBM could enhance survival signalling and tumour progression. The IHC results indicated a weak PIK3CA/AKT/mammalian Target of Rapamycin (mTOR) signalling with the canonical PIK3CA E545K mutation. Moreover, NF1 alterations are negatively linked to GBM patients’ survival and are enriched in the mesenchymal subgroup of GBM. Previous studies in NF1 and Transformation-Related Protein 53 (TRP 53) mutant mice indicated that loss of NF1 in combination with loss of the gene encoding the p53 protein predisposes mice to astrocytoma and GBM, indicating that NF1 likely can play a causal role in GBM tumorigenesis [26-31].
Furthermore, the analysis of IMvigor210 cohort supported that GBM patients in the low-risk group were more likely to make significant responses to anti-PD-L1 immuno-therapy. Emerging studies reported that PD-L1 level within the GBM corresponds to tumour aggressiveness and malignancy level resulting in the risk of immune evasion. PD-L1 highly expressed in most GBM patients and some of tumour tissues detected the PD-L1 level more to 61%. Clinical reports of anti-PD-L1 as monotherapy evaluated the efficacy of treatments. Thus, the establishment of predictive biomarkers for checkpoint immunotherapy was vital in maximizing the therapeutic benefit [32-35].
More in-depth clinical factors like different tumour regions both the core of the tumour or the invasive margin, should be incorporated to prediction models for improvement of accuracy. Currently, our comprehensive and systematic evaluation of the cellular and molecular and genetic factors linked with 6-genes signature has yielded certain insights, which might shed light on how tumours respond to immunotherapies and guide the development of novel clinical drug combination strategies.
The construction of 6-genes prognostic signature and verification of significant association between prognostic signature and clinical survival prognosis, genetic alteration, immune efficacy might demonstrate its potential success of immune checkpoint blockade in combination with PIK3CA and NF1 signature inducing tumour immune sensitivity.
PIK3CA and NF1 signature model might be determined to be an independent and robust prognostic biomarker. In brief, this PIK3CA and NF1 signature exhibited its good and reliable ability in prognosis prediction and would promote personalized treatment and precision medicine. Consistent with our analysis about study individual mutations and their role in cancer immunotherapy, which might provide a novel insight on the mechanism of PIK3CA and NF1 mutation in the GBM.
We thank Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University and Department of Orthopaedics and the first clinical medical college of Guangzhou University of Chinese and Department of Orthopaedics, Jiangmen Hospital of Traditional Chinese Medicine Affiliated to Jinan University.
Conception and design: Qinbiao Chen, Min Luo and Wenjing Lin. Acquisition of data: Qinbiao Chen, Cheng Zhong and Anmin Liu. Curation of data: Qinbiao Chen and Lin Xie. Drafting the article: Qinbiao Chen and Linlin Lu. Critically revising the article: All authors. Study supervision: All authors. All authors read and approved the final manuscript.
The authors certify that all the original data in this research could be obtained from public database. Other data used to support the findings of this study are included within the supplementary information files. All the raw data of this study are available from the first author or corresponding author upon request.
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
Citation: Xie L, Liu A, Chen Q, Luo M, Lu L, Lin W, et al (2024) Analysis of Integrating PIK3CA and NF1 Mutations into Glioblastoma Multiforme Prognostication and Immunotherapy Treatment Strategy. J Clin Chem Lab Med. 7:287.
Received: 27-May-2024, Manuscript No. JCCLM-24-30806; Editor assigned: 29-May-2024, Pre QC No. JCCLM-24-30806 (PQ); Reviewed: 14-Jun-2024, QC No. JCCLM-24-30806; Revised: 21-Jun-2024, Manuscript No. JCCLM-24-30806 (R); Published: 28-Jun-2024 , DOI: 10.35248/2736- 6588.24.7.287
Copyright: © 2024 Xie L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.