Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research Article - (2024)Volume 12, Issue 1
Antifungal human gut lactobacilli with potential probiotic properties is a promising and adaptable approach in gut health balance. Antifungal property of lactobacilli can be favourable in case of mycotic human gut infections like oesophageal candidiasis, ulcers, mycotoxicosis etc. In vitro probiotic characteristics of human gut lactobacilli with antifungal activity against Aflatoxin B1 (AFB1) producing Aspergillus flavus Microbial Type Culture Collection and Gene Bank (MTCC) 2798 was evaluated. Ten lactobacilli isolates from human faecal samples were isolated. The antifungal property of the lactobacilli against Aspergillus flavus MTCC 2798 were studied by agar overlay method. The isolates identified L1, L2, L3, L4, L5, L6, L7, L8 as Limoslactobacillus fermentum, L9 as Lactiplantibacillus plantarum and L 10 as Ligilactobacillus salivarius. They were studied for in vitro probiotic characteristics. L. fermentum L2 MW600479 showed β-hemolytic activity. Virulence genes, gelE, hyl, asa1, efaA, esp, cylA, ace were studied and their absence confirmed the safety status of the lactobacilli isolates. Simulated gastrointestinal tolerance was highest for L. fermentum L8 MW485761 (93.55%). In vitro cell adhesion studies showed maximum auto aggregation for L. fermentum L4 MW600464 (78%), maximum co-aggregation with E. coli O157:H7 MT912681 for L. fermentum L1MW600457 (70.65%) and maximum cell surface hydrophobicity to chloroform for L. fermentum L5 MW600493 (80.35%). Bacteriocin genes Plantaricin (Pln) EF, Pln C were detected in all isolates. 2, 2-Diphenyl-1-Picrylhydrazyl (DPPH) radical scavenging assay showed strong scavenging activity for L. fermentum L8 MW485761 (74.8%). The human gut lactobacilli isolates showed antifungal activity against Aspergillus flavus MTCC 2798. Nine lactobacilli sps showed satisfying in vitro probiotic properties, with bacteriocin production and absence of virulent genes, can further be used as promising candidates for probiotic formulation for gut health.
Antifungal; Human gut; Lactobacilli; Probiotic; Bacteriocin
LAB: Lactic Acid Bacteria; GRAS: Generally Regarded As Safe; OD: Optical Density; LB: Luria Bertani; CFS: Cell Free Supernatant; AFB1: Aflatoxin B1; PDA: Potato Dextrose Agar; MRS: de Man Rogosa and Sharpe; SDA: Sabouraud Dextrose Agar; DNase: Deoxy Ribonuclease; SGJ: Simulated Gastric Juice; SPJ- Simulated Pancreatic Juice; SSJ: Simulated Salivary Juice; PBS: Phosphate Buffered Saline; MHA: Mueller Hinton Agar; CFU: Colony Forming Unit; DPPH: 2,2-diphenyl-1-picrylhydrazyl; PCR: Polymerase Chain Reaction
Food contamination by aflatoxin producing fungi like Aspergillus flavus and Aspergillus parasiticus during storage especially in warm conditions is a major concern in the food industries. Dietary aflatoxin exposure is documented in tropical climate zones such as sub-Saharan Africa and Southeast Asia. Aflatoxins cause carcinogenic, teratogenic and growth stunting effects, hence considered a major food safety concern [1]. Various physical and chemical methods for prevention of fungal and aflatoxin contamination are currently used, but are non-biodegradable and must be applied to products via heat, ionizing radiation or pesticide and fungicide spraying and it causes both sensory changes and irreversible environmental damage [2]. Probiotic bacteria are potential means to fight Aspergilli and reduce the fungal growth and availability of aflatoxin. Lactobacilli cultures from native fermented foods with probiotic attributes and mycotoxin binding character are satisfactory in decontaminating mycotoxins in food [3]. Probiotics are live microorganisms which when administered in adequate quantity provide health benefits to the hosts Food Agricultural Organization (FAO/WHO.2001). Lactic Acid Bacteria (LAB) produce bioactive compounds such as reuterin, cyclic peptides, fatty acids, etc. with antifungal properties. The genus Lactobacilli are gram-positive, catalase negative, oxidase negative, non-spore forming rods of LAB group [4]. Biocontrol of fungus and mycotoxins using Generally Regarded As Safe (GRAS) organisms like LAB can overcome the harm mycotoxin cause to humans.
Human gut microbiota balance overall health; play important roles in mammalian homeostasis, providing essential nutrients, metabolizing dietary fibre into short chain fatty acids and ensuring proper development of the immune system. Therefore, the gut microbiota is considered a crucial factor for proper early life development and lifelong health [5]. In the current situation of Coronavirus Disease-19 (COVID-19), patients have reported intestinal microbial dysbiosis with decreased microbiota such as Lactobacilli and Bifidobacterium; suggesting the need to assess the nutritional and gastrointestinal function of each individual [6]. Lactobacilli have been used as probiotics for disease treatment, prevention of infections; health restoration and maintenance [7]. For a probiotic organism, the efficacy and safety aspects have to be assessed with respect to the criteria for their selection. For safety assessment, tests like hemolytic activity, gelatin hydrolysis and DNase test have to be performed [8]. The probiotic strains should not carry genes for transmissible antibiotic resistance, survive in the gastrointestinal tract, adhere to the human intestinal mucosa; protect it from pathogens by aggregation, co-aggregation and should show antagonistic activity against gastrointestinal pathogens. Probiotic strains which are a part of the human microbiota with Generally Recognized as Safe (GRAS) status are promising candidates [9]. Probiotic potential of such strains can be used for health benefits. A probiotic formulation from human gut with antifungal and probiotic attributes is a novel approach. Our study focuses on the evaluation of in vitro probiotic characteristics of all the 10 human gut Lactobacilli with antifungal activity against AFB1 producing Aspergillus flavus MTCC 2798.
Sample collection
Fecal samples from 32 healthy human volunteers of different age groups, 8 months to 75 years were collected. Early morning fecal samples were collected in sterile collection containers, transported with ice and plated within 2 h of sample collection. They had no history of gastrointestinal disorder and had not received antibiotics 3 months prior to the study. An informed consent form was signed by the healthy volunteers before sample collection.
Aflatoxin B1 producing fungi
Aspergillus flavus MTCC 2798 (produce AFB1) was procured from IMTECH, Chandigarh; grown and maintained on Potato Dextrose Agar (PDA), himedia at room temperature (25-30oC) for 10 days until sporulation occurs, stored as oil overlay culture at 4oC.
Other microorganisms
E. coli MTCC 1610, Klebsiella pneumonia MTCC 432, Salmonella typhi MTCC 733 and Staphylococcus aureus MTCC 96 from IMTECH, Chandigarh, Salmonella paratyphi (a clinical isolate) and E. coli O157:H7 MT912681 ( Dr. Sudha K, St. Peter’s College, Kolencherry, Kerala) , Bacillus subtilis (soil), Enterococcus faecalis ATCC 29212.
Isolation of human gut Lactobacilli
Faecal samples collected as mentioned above were serially diluted in sterile peptone water (0.1% w/v), 100 µl were spread plated onto de Man Rogosa and Sharpe agar (MRS) from himedia and incubated at 37°C for 48 h under partial anaerobic condition in a candle jar [10]. Colonies with different morphology were subjected to preliminary screening for lactobacilli with gram staining, catalase and oxidase reaction. Glycerol stocks were prepared with 50% glycerol and stored at -80ºC.
Screening for antifungal Lactobacilli
Preparation of fungal inoculum: Aspergillus flavus MTCC 2798 were grown on PDA slant at room temperature (25-30oC) for 7-10 days or until sporulation occurs. The spores were dispersed in 4 ml phosphate buffer and adjusted to get an absorbance of 0.35 at 600 nm in an ELICO double beam SL 210 ultraviolet visible spectrophotometer [11].
Detection of the antifungal activity of isolates by agar overlay method: Overnight grown cultures of Lactobacilli isolates with Optical Density (OD) 1 ± 0.2 were patched in 2 cm length onto MRS agar plates and were incubated at 37°C for 48 h [12]. These culture plates were overlayed with 30 ml soft agar (0.75% agar) preparation of Sabouraud Dextrose Agar (SDA), himedia containing 1 ml inoculum (0.35 measured at 600 nm/105 spores/ ml) of fungal spores. The plates were incubated at room temperature for 2 days and examined for clear zones of inhibition around the bacterial growth. The zone of inhibition of Aspergillus flavus MTCC 2798 was estimated using a semi-quantitative scale: (-) no inhibition, (+) minimal inhibition, (++) partial inhibition and (+++) total inhibition. Plates containing fungal spore inoculums only were used as a control.
Deoxyribonucleic acid isolation, blast identification and phylogenetic analysis: Genomic Deoxy Ribonucleic Acid (DNA) from the ten Lactobacilli with antifungal activity was isolated using Mag Genome express DNA bacteria mini kit (Maggenome Pvt Ltd.). Polymerase Chain Reaction (PCR) amplification was done using universal bacterial identification primers 16 SF (5’-AGA GTT TGA TCM TGG CTC-3’) and 16 SR (5’-AAG GAG GTG WTC CAR CC-3’), [13]. The PCR conditions used were; an initial denaturation step for 5 min at 95°C, 35 cycles of denaturation for 1 minute at 95°C, annealing for 1 minute at 55°C, extension for 5 min at 72°C and final extension for 7 min at 72°C in Thermal cycler Biochemicals-Radiochemicals (BIO-RAD) T100 PCR machine [14]. Amplified DNA was sequenced using big dye terminator sequence reaction ready mix (Applied bio system). The Smaller Subunit ribosomal Ribonucleic Acid (16SrRNA) sequence data were checked for similarity analysis with National Centre for Biotechnology Information- Basic Local Alignment Search Tool (NCBI-BLAST) program and submitted to the GenBank sequence database; accession numbers were obtained. The phylogenetic analysis of the isolates obtained was carried out with Molecular Evolutionary Genetics Analysis (MEGA) 11 using the neighbour-joining method with 1000 bootstrap replicates and a phylogenetic tree was generated.
In vitro screening of isolates for probiotic characteristics
Safety assessment tests: Safety assessment tests were performed for the ten Lactobacilli isolates. Hemolytic activity was screened with sheep blood agar plates [15]. DNase test was performed with Deoxy Ribonuclease (DNase) agar [16]. S. aureus MTCC 96 was used as positive control for the above tests. Gelatin hydrolysis was performed and B. subtilis was used as positive control [17] . Antibiotic sensitivity test was done on MRS plates [18]. Antibiotic discs of Chloramphenicol (30 mcg), Streptomycin (10 mcg), Penicillin G (10 mcg), Sparfloxacin (5 mcg), Ampicillin (10 mcg), Tetracycline (30 mcg), Pefloxacine (5 mcg), Ciprofloxacin (5 mcg), Gentamycin (10 mcg), Erythromycin (15 mcg), Teicoplanin (30 mcg), Carbenicillin (100 mcg), Vancomycin (30 mcg) and Kanamycin (30 mcg) (HI-MEDIA) were used. The zones of inhibition were observed and diameters were compared with the standard chart.
Detection of virulence genes: Virulence genes gel E, hyl, asa1, efaA, esp, cytA, ace was studied [19]. E. faecalis ATCC 29212 was used as positive control. Bacterial genomic DNA isolated using Mag genome DNA isolation kit (Mag genome Pvt Ltd.) were used to detect the presence of virulence genes using gene-specific primers (Table 1). The reaction mixture consists of 12.5 µl PCR master mix (Thermo Scientific), 1 µl of each of two primers (Sigma-Aldrich), 1 µl template DNA of the test bacteria and 9.5 µl deionized water (Sigma). Following temperature program: Initial denaturation at 94°C for 5 min, 30 cycles of 94°C for 45 s, 50°C –64°C (according to the annealing temperature for the individual primers) for 45 s, 72°C for 75 s and a final extension step at 72°C for 8 min were used (Table 1). 2% agarose gels were used to separate PCR products.
| Virulent genes | Sequences (5'→3') | Size (bp) | PCR conditions |
|---|---|---|---|
| Gelatinase (Gel E) | F: TATGACAATGCTTTTTGGGAT | 213 | (94°C 1 min, 56°C 1 min, 72°C 1 min) × 30 1 |
| R: AGATGCACCCGAAATAATATA | |||
| Hyaluronidase (Hyl) | F: ACAGAAGAGCTGCAGGAAATG | 276 | |
| R: GACTGACGTCCAAGTTTCCAA | |||
| Aggregation substance (asa1) | F: GCACGCTATTACGAACTATGA | 375 | |
| R: TAAGAAAGAACATCACCACGA | |||
| Endocarditis antigen | F: GCCAATTGGGACAGACCCTC | 688 | |
| R: CGCCTTCTGTTCCTTCTTTGGC | |||
| Enterococcal surface protein (esp) | F: AGATTTCATCTTTGATTCTTG | 510 | |
| R: AATTGATTCTTTAGCATCTGG | |||
| Cytolysin A (cytA) | F: ACTCGGGGATTGATAGGC | 688 | |
| R: GCTGCTAAAGCTGCGCTT | |||
| Adhesion of collagen (ace) | F: GAATTGAGCAAAAGTTCAATCG | 350 | |
| R: GTCTGTCTTTTCACTTGTTTC |
Note: PCR: Polymerase Chain Reaction.
Table 1: Virulence genes, their primers and PCR conditions.
Tolerance test: MRS agar plates of pH 2, 3, 4 and 10 were used to test acid tolerance. The growth was observed after 24 h and 48 h of anaerobic incubation at 37oC [20]. Bile tolerance were tested by MRS broth with sodium taurocholate (0.1%, 0.2%, 0.3%, 1%, 1.5%, 2% and 3%) and incubated at 37°C [20]. From each concentration, a loopful of broth were patched onto MRS agar plates at 0 h, 1 h, 2 h, 3 h, 24 h and 48 h, observed for growth after incubation at 37°C for 24 h. NaCl tolerance was screened with MRS broth with 2%, 3%, 4%, 6.5% and 10% NaCl [21]. Growth was observed at 0 h, 1 h, 2 h, 3 h, 24 h and 48 h as with bile tolerance. Temperature tolerance was screened with MRS plates containing isolates incubated at temperature: 10°C, 25°C, 40°C and 60°C, under anaerobic conditions [20]. The plates were observed for the growth at 24 h up to 5 days. 0.4%, 0.5% and 0.6% of phenol in MRS broth tubes was incubated at 37°C for 24 h under anaerobic conditions to study phenol tolerance. A loopful of inoculum was patched onto MRS agar plates. Plates were incubated at 37°C for 24 h anaerobically and observed for growth [22].
Simulated gastrointestinal tolerance test: Tolerance to gastrointestinal conditions of Lactobacilli isolates were evaluated [23]. Briefly 3 ml of overnight bacterial suspensions with OD (1 ± 0.2) were centrifuged at 5,000 g for 15 min at 4oC, washed and suspended in 3 ml PBS. The pellets were diluted 1:1 (3 ml pellet suspension: 3 ml SSJ (sterile electrolyte solution containing 6.2 g/l NaCl, 2.2 g/l KCl, 0.22 g/l CaCl2 and 1.2 g/l NaHCO3, in which lysozyme (Sigma Aldrich) had a final concentration of 100 mg/L; pH 6.9) in Simulated Salivary Juice (SSJ), incubated for 5 min at 37oC. The sample was diluted 3:5 (3 ml SSJ mix: 5 ml SGJ), with Simulated Gastric Juice (SGJ) containing (6.2 g/l NaCl, 2.2 g/l KCl, 0.22 g/l CaCl2 and 1.2 g/l NaHCO3 at pH 2.5 and with 0.3% pepsin (himedia), incubated for 1 h at 37oC. After incubation, 1 ml of the sample was serially diluted and plated onto MRS agar. The remaining sample was diluted 1:4 (1 ml SGJ mix: 4 ml SPJ) in Simulated Pancreatic Juice (SPJ) (consisting of 6.4 g/l NaHCO3, 0.239 g/l KCl, 1.28 g/l NaCl, 0.5% bile salts and 0.1% pancreatin (himedia), pH 7.2; incubated for 3 h at 37oC. After 2 and 3 h of incubation time, 1 ml of sample was serially diluted and 100 µl of the sample from 10-5 dilution was plated on MRS agar. Aliquots of overnight inoculums were tested to determine the CFU/ml at the initial time point (t0). Survival capacity was calculated as the percentage of 1– [(log CFU/ml t0- log CFU/ml SPJ 3 h)/log CFU/ml t0], where CFU/ ml SPJ 3 h represented the total viable counts (CFU/ ml) for each isolate at the final time point of incubation in SPJ and CFU/ ml t0 represented the total viable counts at t0.
In vitro adhesion studies
Auto aggregation: Lactobacilli isolates were inoculated in MRS broth, while pathogenic strains were inoculated in Luria-Bertani broth (LB) and incubated overnight at 37°C. Bacterial cells were pelleted in a refrigerated centrifuge (Eppendorf, 5804 R) at 6000 g for 10 min. The pellets were washed twice with Phosphate Buffered Saline (PBS) pH 7.0 after washing, the pellets were resuspended in PBS and Optical density (OD) was adjusted to 0.60 ± 0.02 at 600 nm. From this, 4 ml of this suspension was transferred to a sterile tube and incubated at 37°C without agitation for 24 h. 0.1 ml of the cell suspension was transferred from the surface to another tube containing 3.9 ml of PBS and the OD was measured at 600 nm at 0th h and 24 h of incubation in a ELICO double beam sl 210 uv visible spectrophotometer [24]. The OD values at selected hours of incubation were recorded and corresponding auto aggregation % was calculated using the formula:
Auto aggregation % = ([A1-A2])/ (A1) × 100
Where, A1 is the OD at 0th h and A2 is the OD measured after incubation at 24 h.
Co-aggregation: Cell suspensions of the isolates and the pathogenic strain (E. coli O157: H7 MT912681) were prepared as mentioned above. 2 ml of the Lactobacilli cell suspension was mixed with 2 ml of E. coli O157:H7 MT912681 suspension and vortexed for 10 s, incubated at 37oC. 0.1 ml of the undisturbed upper suspension was transferred to another tube with 3.9 ml of PBS and the OD was measured at 600 nm at 24 h in an ELICO double beam Sl 210 UV visible spectrophotometer. OD values were recorded for respective hours and the co-aggregation percentage was calculated using the equation of [25].
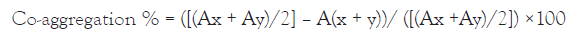
Where, A stands for absorbance; x and y represent each of the two strains in the control tubes and (x + y) that of the mixture.
Cell surface hydrophobicity: The hydrophobicity of Lactobacilli isolates was evaluated by the microbial adhesion to chloroform, an electron-acceptor solvent with slight modifications [26]. Briefly, the cell suspensions of the isolates were prepared as mentioned in the case of auto aggregation. 6 ml of the cell suspensions of each isolates in PBS was mixed with 2 ml of the solvent and vortexed for 90 s. The mixture was allowed to stand for 1 h at room temperature to ensure the complete separation of the two phases after which the aqueous phase was carefully collected and the OD was measured at 660 nm in ELICO double beam SL 210 UV visible spectrophotometer. The percentage of cells binding to the solvent was subsequently calculated using the equation:
Hydrophobicity % = ((OD before mixing – OD after mixing))/ (OD before mixing) ×100
Other probiotic characterisation tests
Antibacterial activity: Modified cross-streak method (1987) was used to study the antibacterial activity of the isolates [27].A through (1-inch-wide) was cut vertically in the middle of Mueller Hinton Agar (MHA) into which MRS media was poured in for the growth of Lactobacilli isolates. Lactobacilli isolates were patched onto the MRS agar portion of MHA plates and incubated anaerobically at 37°C for 48 h. The plates were then cross streaked with pathogenic strains: E. coli O157:H7 MT912681, E. coli MTCC 1610, K. pneumoniae MTCC432, S. typhi MTCC 733, S. paratyphi and S. aureus MTCC 96. The plates were incubated aerobically at 37°C for 24 h and observed for inhibition.
Bile salt hydrolase activity: Lactobacilli oxgall agar containing 0.5% taurodeoxycholic acid sodium salt and 0.037% calcium chloride per 100 ml were used. The isolates were patched onto oxgall agar and incubated anaerobically for 72 h and observed for precipitation around the growth or opaque halos around the colonies.
Bacteriocin production
Qualitative test for bacteriocin production: Spot on lawn method is with slight modification was performed. MRS plates (1.5% agar) were spotted with Lactobacilli isolates, incubated anaerobically overnight at 37°C. To confirm that a bacteriocin like substance was produced, a hole in the agar was punched next to a colony with a toothpick. The hole was filled with 10 μl of bovine serum albumin (control) and proteinase K (each at 20 mg/ml), incubated for 2 h at 30°C. The spotted plates were overlaid with LB agar (1%), seeded with E. coli O157:H7 MT912681 (105 -106 CFU). The plates were incubated anaerobically overnight at 37°C. After incubation for 15 h the plates were examined to judge whether the inhibitory substance was sensitive to proteolysis [28].
Detection of bacteriocin genes: PCR assay was used to determine bacteriocin genes plantaricin EF (plnEF), plantaricin A (plnA), plantaricin C (plnC). Gassericin A (gaaA), plantaricin S (plnS) and lactacin (laf) [29]. PCR specific primers and annealing temperatures are shown (Table 2). PCR conditions of the genes were: An initial denaturation step of 95°C for 5 min, 34 cycles of 95°C for 1 min, extension 72°C for 45 s, followed by a final extension at 72°C for 5 min. The amplified products were visualized by electrophoresis in 2% agarose gels stained with ethidium bromide. .
| Bacteriocin genes | Sequences (5'→3') | Size (bp) | PCR condition |
|---|---|---|---|
| Plantaricin EF (plnEF) | F: GGCATAGTTAAAATTCCCCCC | 428 bp | (94°C 1 min, 55°C 30 s , 72°C 30 s) × 35 |
| R: CAGGTTGCCGCAAAAAAAG | |||
| Plantaricin A (plnA) | F: GTACAGTACTAATGGGAG | 450 bp | |
| R: CTTACGCCATCTATACG | |||
| Plantaricin C (plnC) | F: GGTGGCGACAGGAGATTTAC | 353 bp | |
| R: AGAAACGCGTTCCGATTTTA | |||
| Gassericin A (gaaA) | F: GAACAGGTGCACTAATCGGT | 800 bp | 94°C 1 min, 62°C 45 s , 72°C 30 s) × 35 |
| R: CAGCTAAGTTAGAAGGGGCT | |||
| Plantaricin S (plnS) | F: GCCTTACCAGCGTAATGCCC | 320 bp | 94°C 1 min, 62°C 1 min , 72°C 1 min) × 35 |
| R: CTGGTGATGCAATCGTTAGTT | |||
| Lactacin (laf) | F: AGTCGTTGTTGGTGGAAGAAAT | 184 bp | |
| R: TCTTATCTTGCCAAAACCACCT |
Note: PCR: Polymerase Chain Reaction.
Table 2: Bacteriocin genes, their primers and PCR conditions.
Antioxidant activity assay by DPPH Radical Scavenging Method: Cell Free Supernatant (CFS) of Lactobacilli were prepared by centrifuging the overnight culture at 10,000 g for 10 minutes, at 4°C. The CFS of each Lactobacilli was adjusted to pH 7.0 with 1 M NaOH. 1 ml CFS of each sample and 1 ml of freshly prepared 2,2-Diphenyl-1-Picrylhydrazyl (DPPH) solution (0.2 mM in 100% methanol) was mixed well and left in the dark at room temperature for 30 min. The blank sample contained deionized water along with the DPPH solution. Following incubation, read OD at 517 nm in ELICO double beam SL 210 UV visible spectrophotometer [30]. The percentage of radical scavenging activity was calculated using the calculation [31].
(AB control-AB sample)/ABcontrol×100
Where, AB is the absorbance at 517 nm.
Primary characterization of isolates
From the fecal samples collected from 32 healthy volunteers, 433 lactic acid bacteria were isolated.175 colonies which were small, off- white to cream colour, having round/spindle shape, Gram positive rods suggestive of lactobacilli were sub-cultured. The colonies were further screened to identify Lactobacilli. Catalase and oxidase negative, gram positive rods were considered as lactobacilli and were studied for their antifungal activity.
Screening for antifungal Lactobacilli
Out of 175 isolates, 10 of them showed consistent antifungal activity against Asp. flavus MTCC 2798 in the agar overlay assay. The isolates L. fermentum L1, L3, L4, L5, L6, L7, L8; L. plantarum L9 and L. salivarius L10 showed strong inhibitory activity (large clear zone around the bacterial patch) and L. fermentum L2 showed moderate inhibitory activity after 48 h of incubation (Figure 1). Earlier reports suggest that pH reduction [32]. Due to the production of organic acids like lactic acids and antagonistic compounds such as hydrogen peroxide; phenolic compounds produced by lactobacilli can inhibit the growth of fungal hyphae [33]. L. delbrueckii UFV H2b20 was reported to have inhibited the mycelial growth of aflatoxin producing Asp. parasiticus IMI 242695 [34].

Figure 1: Antifungal effect of lactobacilli isolates against Aspergillus flavus Microbial Type Culture Collection (MTCC) 2798 using agar overlay method. (A): Control plate; (B): Isolate with no inhibition; (L1-L10): Isolates showing inhibition.
Deoxyribonucleic acid isolation, blast identification of bacterial isolates, phylogenetic analysis
16 S rDNA sequences (SciGenom) of 10 Lactobacilli showing antifungal activity were sequenced and identified by NCBI-BLAST program. The sequences were submitted in NCBI Gen Bank, accession numbers were obtained (Table 3). Phylogenetic tree were constructed based on neighbor-joining analysis of 16 S rRNA gene (Figure 2). L. fermentum L1, L2, L3, L4, L5, L6, L7, L8 strains were categorised in L. fermentum strain ATCC 14931 M58819 cluster with 100% bootstrap cluster similarity. L. salivarius AL4 L10 showed 100% bootstrap cluster similarity with L. salivarius JCM 1231 AB370881 cluster. L. plantarum AL1 L9 showed 99%bootstrap cluster similarity to L. plantarum JCM 1149 HM162417 cluster. The utilization of LABs as probiotics makes their identification and characterization important. Molecular tools such as nucleotide sequencing, real time PCR and phylogenetic analysis are more powerful and accurate for differentiation and characterization of LABs than traditional methods (Sharifpour, et al., (2016)) [35].
| Isolate | Lactobacillus strains identified | Gene Bank Accession no. |
|---|---|---|
| L1 | Limosilactobacillus fermentum | MW600457 |
| L2 | Limosilactobacillus fermentum | MW600479 |
| L3 | Limosilactobacillus fermentum | MW600480 |
| L4 | Limosilactobacillus fermentum | MW600464 |
| L5 | Limosilactobacillus fermentum | MW600493 |
| L6 | Limosilactobacillus fermentum | MW600495 |
| L7 | Limosilactobacillus fermentum | MW600496 |
| L8 | Limosilactobacillus fermentum | MW485761 |
| L9 | Lactiplantibacillus plantarum | MW485746 |
| L10 | Ligilactobacillus salivarius | MW600498 |
Note: NCBI: National Center for Biotechnology Information; BLAST: Basic Local Alignment Search Tool.
Table 3: Molecular identification of the lactobacilli isolates by NCBI -BLAST.
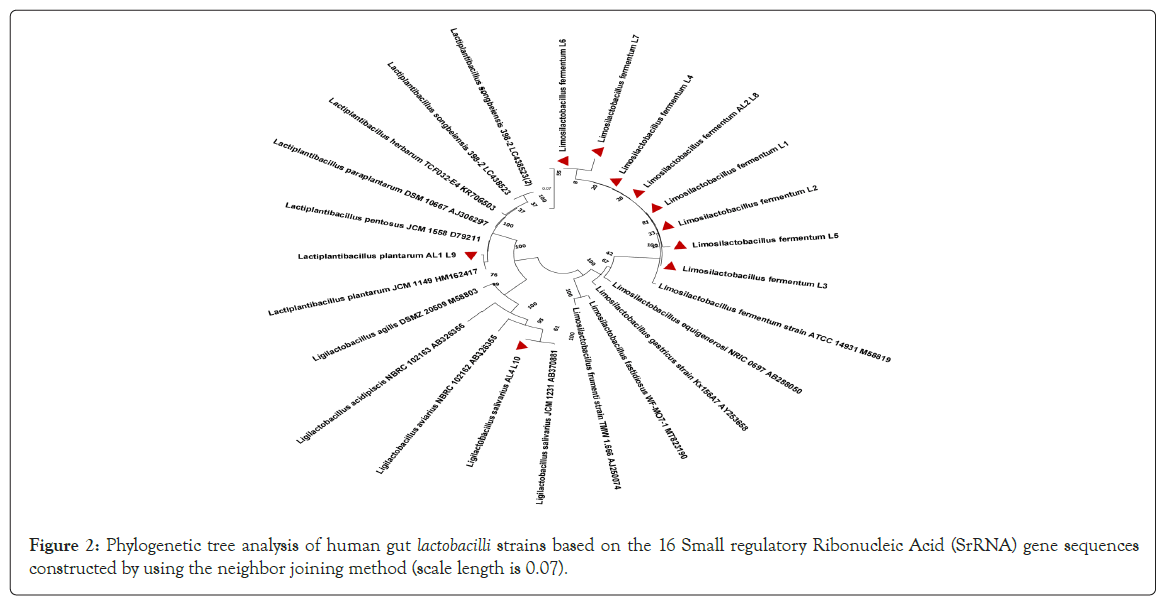
Figure 2: Phylogenetic tree analysis of human gut lactobacilli strains based on the 16 Small regulatory Ribonucleic Acid (SrRNA) gene sequences constructed by using the neighbor joining method (scale length is 0.07).
In vitro screening of Isolates for probiotic characteristics
Potential probiotic Lactobacilli were selected on the basis of functional and safety standards. In the present study, ten lactobacilli of human gut origin with antifungal activity were tested for their in vitro probiotic properties.
Safety assessment test: According to the Food and Agriculture Organization (FAO)/ World Health Organisation (WHO) 2002 working group guidelines for the evaluation of probiotics in food, the study of hemolytic characteristics of probiotic bacteria is significant in assuring safety for human use. Hemolysin is an exotoxin that attacks blood cell membranes and causes cell rupture. Lactobacilli sps. is α-hemolytic microorganisms [36,37]. Some Lactobacilli species of human gut origin exhibit significant β – hemolysis. L. fermentum MW600479 (L2) showed β-hemolysis on sheep blood agar. Hemolytic character of Lactobacilli is considered a virulent trait and hence L. fermentum MW600479 (L2) was eliminated from the study. Remaining nine Lactobacilli isolates were non hemolytic and regarded as safe (Figure 3). Weckman and Catlin (1957) reported that DNase activity could be used to identify potentially pathogenic species. Gelatinase degrades gelatin and collagen to intensify pathogenesis [38]. All the isolates were DNase negative and hence considered safe. All the nine isolates were negative for gelatinase activity, unlike Bacillus subtilis (positive control) which showed a clear zone in gelatin agar. Strains having no gelatinase activity are safe to be marketed as a probiotic (FAO 2006).
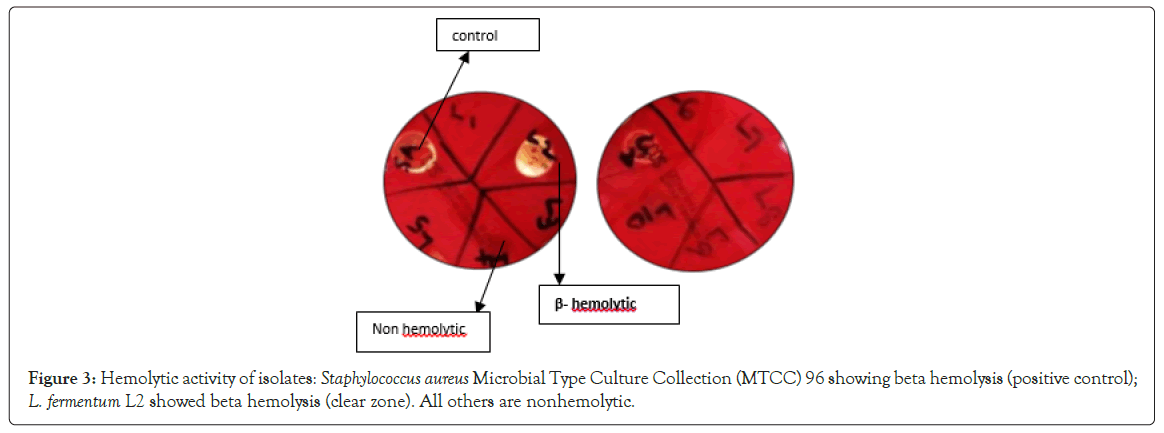
Figure 3: Hemolytic activity of isolates: Staphylococcus aureus Microbial Type Culture Collection (MTCC) 96 showing beta hemolysis (positive control); L. fermentum L2 showed beta hemolysis (clear zone). All others are nonhemolytic.
Antibiotic resistance among the probiotic strains should be innate and non-transferable [39]. Intrinsic antibiotic resistance in probiotic strains could be useful for restoring the gut microbiota after antibiotic treatment. In our study lactobacilli showed resistance for pefloxacine (100%), teicoplanin (90%), vancomycin (90%) and kanamycin (90%). Majority of the Lactobacilli strains are naturally resistant to vancomycin as being chromosomally encoded, the vancomycin resistance is not transferable [40]. Isolates showed a sensitivity pattern to chloramphenicol (100%), tetracycline (100%), erythromycin (90%), gentamycin (80%), penicillin G (70%) and ampicillin (70%) (Figure 4). Tetracycline and erythromycin resistance can be transferred horizontally within the members of same or related species of lactobacilli [41]. All the nine isolates of our study were tetracycline and erythromycin sensitive and thus safe.
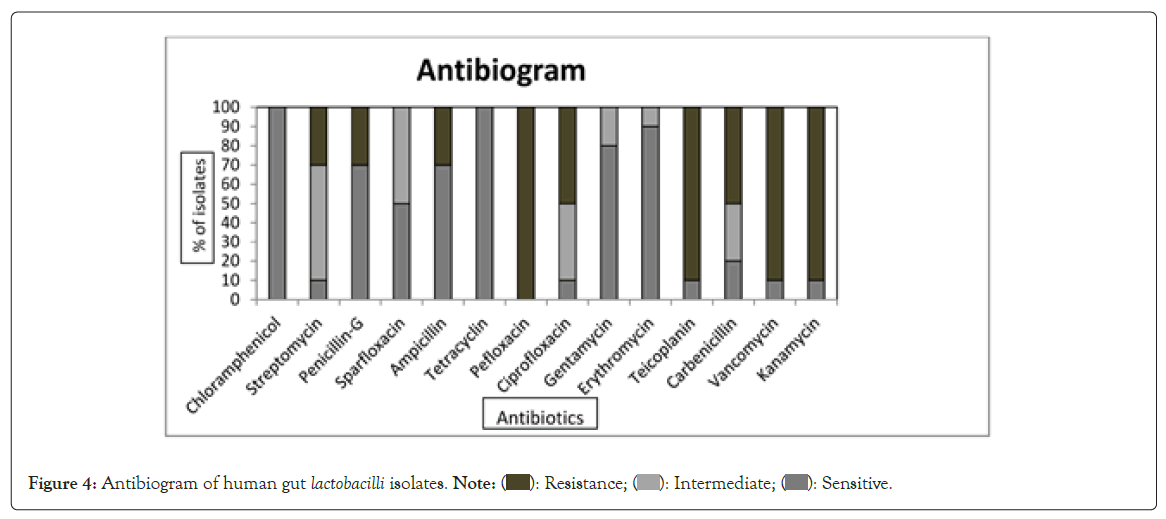
Figure 4: Antibiogram of human gut lactobacilli isolates. Note: (  ): Resistance; (
): Resistance; ( ): Intermediate; (
): Intermediate; (  ): Sensitive.
): Sensitive.
Detection of virulence genes: Human gut lactobacilli isolates showed absence of virulence genes gel E, hyl, asa1, efaA, esp, cytA, ace after PCR. Enterococcus faecalis ATCC 29212 showed the presence of asa1 (aggregation substance) gene (Figure 5). Determination of virulence factors in lactobacilli by molecular and phenotypic procedures are important due to the risk of gene transfer, since these factors are usually encoded by genes located in conjugative plasmids [19]. All 9 isolates have do not carry any virulence genes hence safe for use as probiotics.
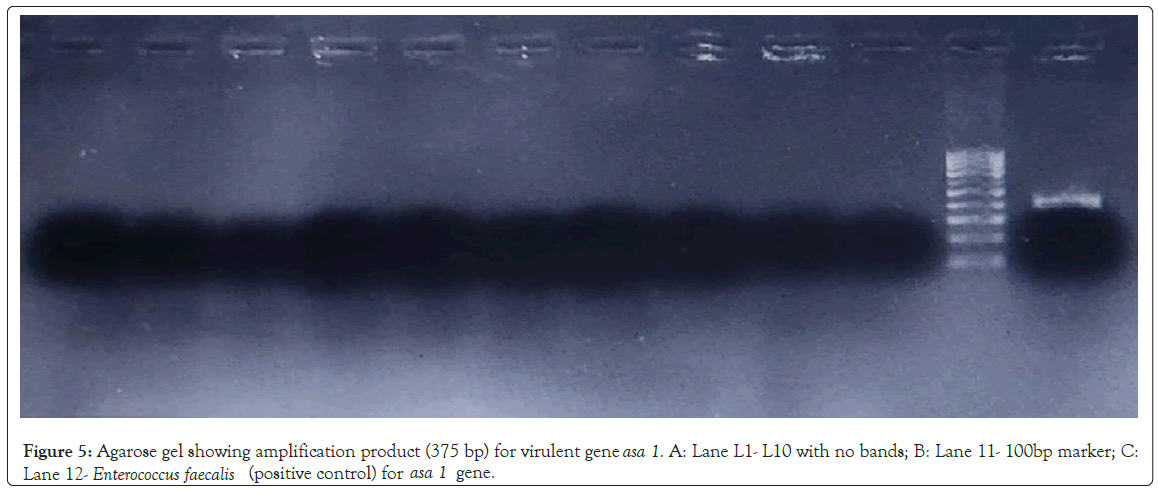
Figure 5: Agarose gel showing amplification product (375 bp) for virulent gene asa 1. A: Lane L1- L10 with no bands; B: Lane 11- 100bp marker; C: Lane 12 - Enterococcus faecalis (positive control) for asa 1 gene.
Tolerance test: Isolates showed good growth in MRS media with pH range 2 to 10 at 24 h and 48 h of incubation. The isolates L. fermentum L1, L3, L4,L5,L6,L7,L8, L. plantarum L9, L. salivarius L10 were able to survive in extreme acidic pH (pH 2 to 4) and L. fermentum L1, L4,L5,L6,L7,L8, L. plantarum L9, L. salivarius L10 were able to survive in basic pH 10, while the growth of isolate L. fermentum L3 was inhibited at pH 10. pH 3 is a standard for studying probiotic strains [42,43]. Reported that Lactobacilli strains exposed to pH 2 survived up to 24 h. Human gut strains can tolerate stomach acidity and intestinal alkalinity and easily adaptable to different pH variations. The acid tolerance of lactic acid bacteria is due to the induction of H+- Adenosine Triphosphatase (ATPase) activity [44]. The variation in the acid tolerance of the selected probiotics is related to the difference in H+-ATPase activity in the probiotics [45].
Lan-Szu and Bart (1999) have substantiated that strains selected as probiotics should tolerate acid and bile for at least 90 min, which is the time needed to cross the gastric barrier. The normal range of bile salt concentration in the human intestinal tract is 0.05%-0.3%. Before selection of probiotic bacteria for human consumption, we must ensure that it resists 0.3% bile concentration [46]. All isolates showed good growth in 0.1%, 0.2%, 0.3% and 1% of bile salt concentration, at 0 h, 1 h, 2 h, 3 h, 24 h and 48 h. In 1.5% and 2% bile salt concentrations L. fermentum L1, L3, L6showed scanty growth at 48 h. In 3% bile salt concentration L. fermentum L1, L4, L5, L6showed scanty growth at 48 h, whereas L. fermentum L3 showed no growth at 48 h. Bile salt tolerance is a prerequisite for colonization of probiotic bacteria to walls of small intestine and also for the proper metabolic activity of bacteria in small intestine.
All the isolates showed confluent growth in different salt concentrations (2%, 3%, 4%, 6.5% and 10%) at 0 h, 1 h, 2 h, 3 h, 24 h and 48 h which indicated their high tolerance to NaCl. When the salt concentration goes high, it allows the bacteria to initiate metabolic activity, thus increasing acid production and helps to ward off non desirable microorganisms. Hence, NaCl tolerance is important for an efficient probiotic strain and also important for preservation techniques [47].
All the isolates were able to grow well at 10°C, 25°C, 37°C and 40°C.The isolates failed to grow at 60°C even after 5 days of incubation, indicating that they are less heat tolerant. Temperature tolerance assures viability with good metabolic activity in the storage system and within the human body. The temperature is a factor which can affect the growth and metabolism of bacteria [48].
Aromatic amino acids derived from dietary or endogenously produced proteins can be deaminated in the gut by bacteria leading to the formation of phenols which have bacteriostatic properties [47]. The isolates (L. fermentum L1, L. fermentum L3–L9) showed growth in 0.4%, 0.5% and 0.6% phenol after 24 h incubation and were tolerant to different phenol concentrations. While L. salivarius L10 showed no growth in 0.5% and 0.6% phenol hence least resistant to phenol. Phenolic compounds can exert a bacteriostatic effect against some lactobacilli strains, thus their tolerance is a desirable property [49].
Simulated gastrointestinal tolerance test: The ability to survive gastrointestinal transit with high viable cell counts is an important probiotic criterion. It also helps in effective colonization by a probiotic strain [50]. Selection of potential probiotic strains with ability to survive in the Gastro Intestinal (GI) tract was performed on the basis of the measurements of strain survival after treatment with simulated salivary juice (pH 6.9), simulated gastric juice (pH 2.5) and simulated intestinal juices (pH 8). All the nine isolates showed good survival percentage with the highest for L8 (93.55%). The survival percentages of the isolates are shown in (Figure 6). The tolerance to stress conditions is highly influenced by the type of bacterial strain, type of growth medium and the incubation conditions [51]. High survival rates showed that the isolates under probiotic studies can retain their metabolically active state and transiently colonize and interact with resident gut bacteria [52].
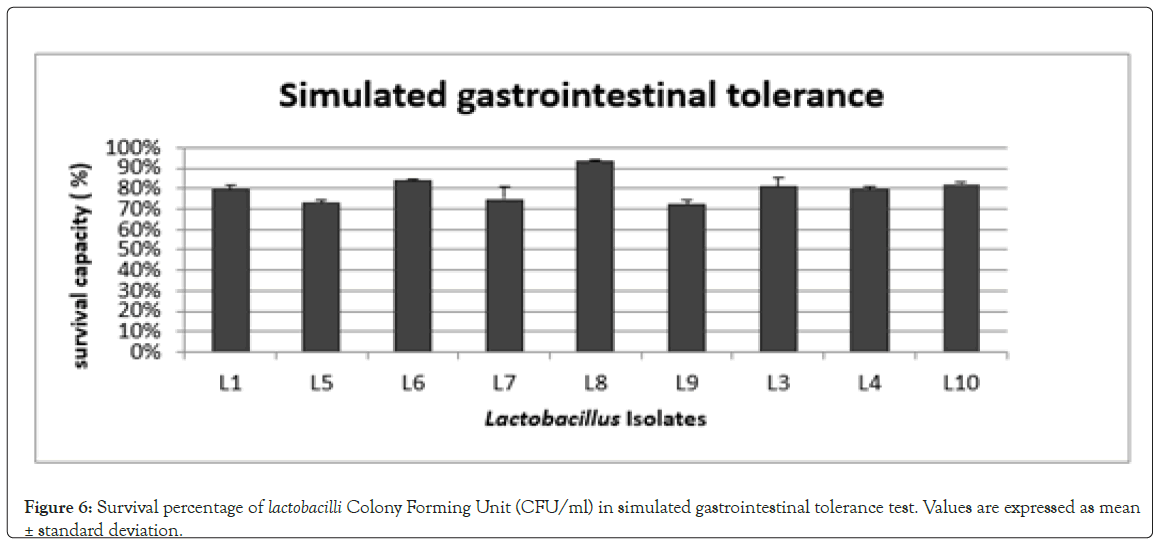
Figure 6: Survival percentage of lactobacilli Colony Forming Unit (CFU/ml) in simulated gastrointestinal tolerance test. Values are expressed as mean ± standard deviation.
In Vitro adhesion studies
Auto aggregation: Auto aggregation is described as the aggregation between microorganisms of the same strain. Auto aggregation ability was expressed as percentage reduction of the absorbance of the bacterial suspension by 24 h. Percentage of auto aggregation ranged from 78% (L. fermentum L4) to 27% (L. fermentum L6) and is shown in (Figure 7). Aggregation in probiotic organisms plays an important role in the formation of biofilms to protect the host from colonisation by pathogens [53,54]. Showed that auto aggregation of Lactobacilli is correlated with their adhesion ability to epithelial cells and mucosal surfaces.
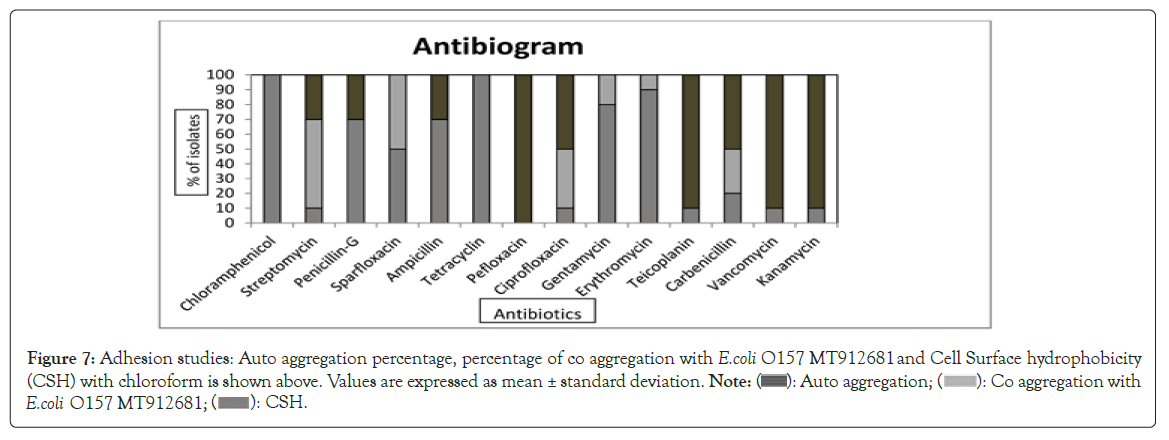
Figure 7: Adhesion studies: Auto aggregation percentage, percentage of co aggregation with E.coli O157 MT912681 and Cell Surface Hydrophobicity (CSH) with chloroform is shown above. Values are expressed as mean ± standard deviation. Note: ( ): Auto aggregation; (
): Auto aggregation; ( ): Co aggregation with E.coli O157 MT912681; (
): Co aggregation with E.coli O157 MT912681; ( ): CSH.
): CSH.
Co aggregation: Co aggregation is the ability of microorganism to interact closely with other species of bacteria. Co aggregation between the native bacteria and pathogens helps to exclude the pathogenic bacteria from their hosts , as the antibacterial substances secreted acts directly on the target pathogen. All the nine isolates were able to co aggregate with the pathogen (E. coli O157:H7 MT912681). L. fermentum L1 had a co-aggregation percentage of 70.65%. L. fermentum L5 showed least co-aggregation ability (5.80%) and is shown in (Figure 7) at 24 h. Co aggregation shows the protective properties of probiotics against pathogens [55]. When comparing the co aggregation studies with antibacterial activity, L. fermentum L1 was most effective [56].
Cell surface hydrophobicity: Cell Surface Hydrophobicity (CSH) is related to adhesive and gut colonization ability [57]. CSH is the ability of the bacteria to partition into hydrocarbons i.e., hexadecane, xylene, toluene, chloroform and reflects the nonspecific adhesion capability related to cell surface characteristics [58,59]. In this study, strong hydrophobicity was shown to chloroform after an incubation period of 1 h. The CSH percentage of the isolates are shown in (Figure 7). L. fermentum L5 exhibited maximum hydrophobicity (80.35%) and L. fermentum L4 exhibited the minimum hydrophobicity (0.68%). The adherence property is due to complex interactions of positive and negative charges between hydrophobic and hydrophilic components of the bacterial surface [60].
Other probiotic characterisation test
Antibacterial activity: Probiotics helps to prevent gastrointestinal disorders by maintaining homeostasis of the gut microbiome, competitively inhibiting the growth of pathogens [61]. L. fermentum L1, L5, L6, L7, L. salivarius L10 partially inhibited E. coli O157:H7 MT912681; L. fermentum L1, L6, L7 inhibited Klebsiella pneumonia MTCC 432; L. fermentum L1, L3, L5, L6, L7 inhibited Salmonella typhi MTCC 73; L. fermentum L5, L6, L. salivarius L10 inhibited Staphylococcus aureus MTCC 96; L. fermentum L1, L3, L5, L6, L7, L. salivarius L10 inhibited Salmonella paratyphi while, L. fermentum L1, L7, L. salivarius L10inhibited E.coli MTCC 1610. L. fermentum L4, L8, L9 showed minimum antibacterial activity while, L. salivarius L10 showed maximum antibacterial activity (Figure 8). The antibacterial activity is due to the production of lactic acid that lower the pH of the medium, production of bacteriocins, diacetyl and hydrogen peroxide formed during lactic acid fermentation or due to the competition for nutrients [20,47,62]. This allows lactobacilli to survive in the presence of the gastrointestinal pathogens [63]. Disparity in the antagonistic activity against different pathogens indicates that probiotic strains are highly pathogen-specific [64].
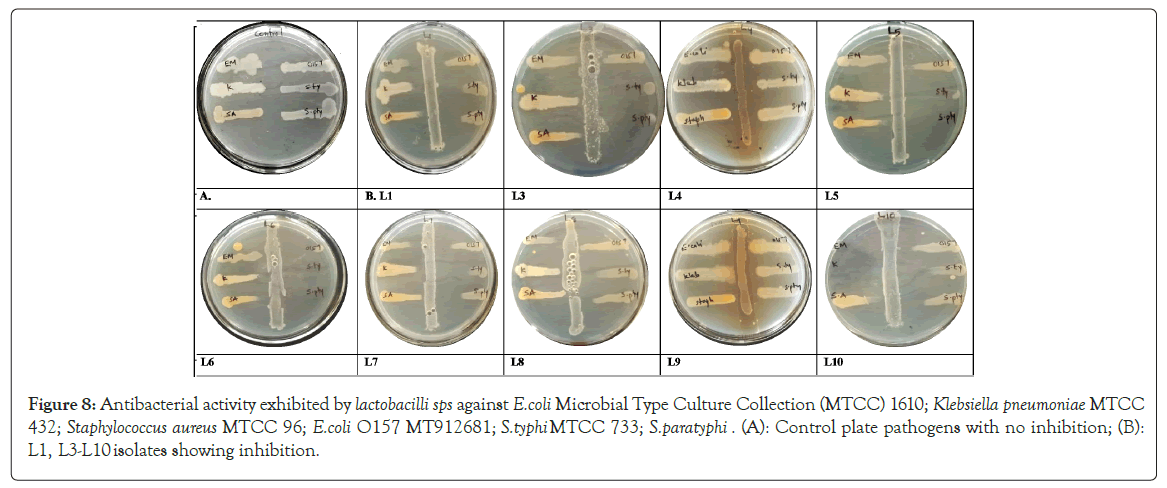
Figure 8: Antibacterial activity exhibited by lactobacilli sps against E.coli Microbial Type Culture Collection (MTCC) 1610; Klebsiella pneumoniae MTCC 432; Staphylococcus aureus MTCC 96; E.coli O157 MT912681; S.typhi MTCC 733; S.paratyphi. (A): Control plate pathogens with no inhibition; (B): L1, L3-L10 isolates showing inhibition.
Bile salt hydrolase activity: The ability of probiotic microorganisms to detoxify bile salt by producing Bile Salt Hydrolysing (BSH) enzyme activity was included among the criteria for probiotic strain selection [65]. It helps the bacteria to thrive in the bile salt environment. The production of opaque granular white colonies with precipitation around colonies or halos indicated bile salt hydrolase activity. Copious amount of deoxycholic acid get precipitated around BSH producing colonies, giving it white granular appearance. Good growth with opacity and precipitation around the colony were observed for L. fermentum L1, L5, L6, L7, L8, L. plantarum L9, L. salivarius L10 indicating BSH activity (Figure 9). L. fermentum L3 and L4 showed no opacity or precipitation around the colony and lack BSH activity (Figure 9). BSH property has been associated with lowering of serum cholesterol level thus BSH activity could control hypercholesterolemia. BSH activity help bile detoxification, gastrointestinal persistence, also allows membrane alterations aiding resistance to bile [66,67].
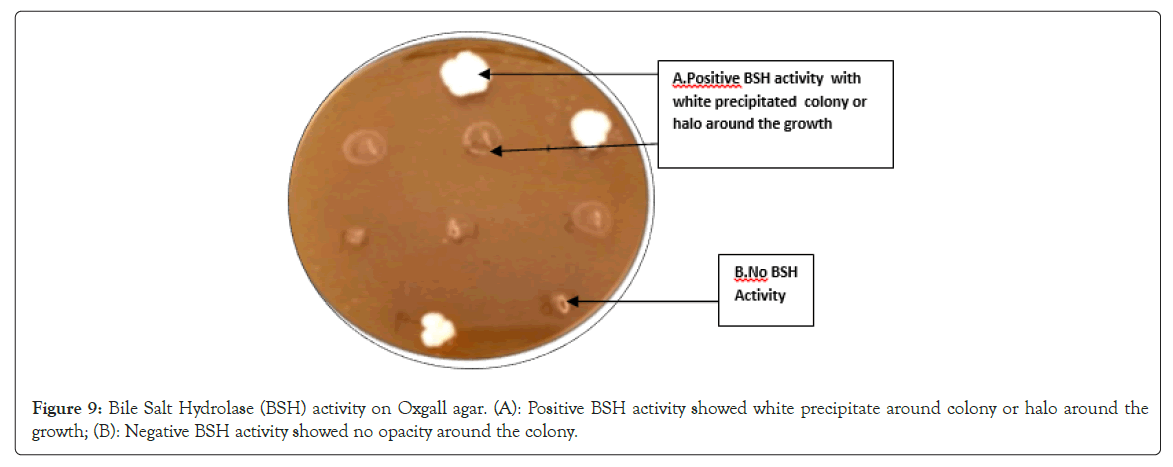
Figure 9: Bile Salt Hydrolase (BSH) activity on Oxgall agar. (A): Positive BSH activity showed white precipitate around colony or halo around the growth; (B): Negative BSH activity showed no opacity around the colony.
Bacteriocin production
Qualitative test: In the spot on lawn method, the inhibition of E. coli O157:H7 MT912681 can be seen as a zone of clearance around the spot for all isolates and the opaque region in the zones are due to inactivation of proteases [68]. Bacteriocins are ribosomally synthesized proteins or peptides produced by bacteria that typically inhibit the growth of closely related species [69]. All the isolates showed opacity in the region where proteinase K was stabbed, the opaque region was produced by degradation of bacteriocin by the proteinase K, thus confirms that the inhibition is due to bacteriocin production (Figure 10). Bacteriocins produced by LAB have attracted special interest because they are GRAS organisms [70].
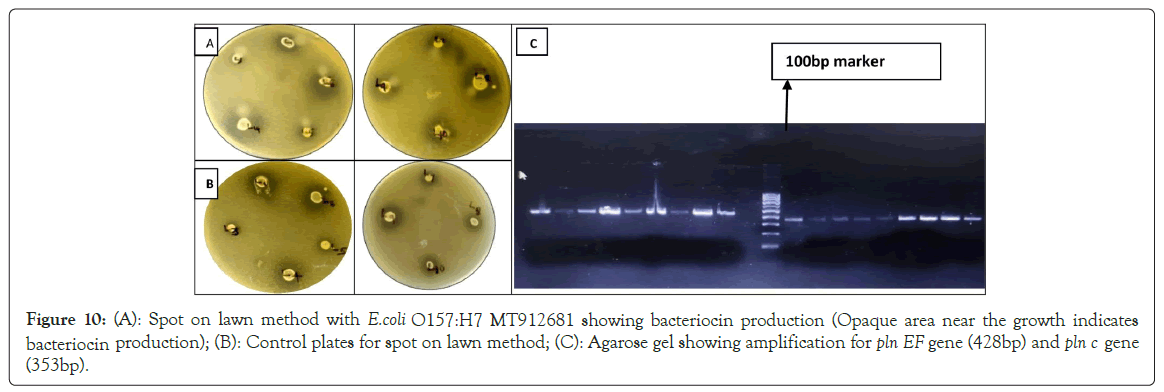
Figure 10: (A): Spot on lawn method with E.coli O157:H7 MT912681 showing bacteriocin production (Opaque area near the growth indicates bacteriocin production); (B): Control plates for spot on lawn method; (C): Agarose gel showing amplification product for pln EF gene (428bp) and pln c gene (353bp).
Detection of the bacteriocin genes: Analysis for rapid screening of bacteriocins-encoding genes by PCR is an alternative method to indicate bacteriocin production. Out of the six bacteriocin genes studied, (pln EF) and (pln C) gene showed amplification. All the isolates showed amplification product for pln EF gene (428 bp) and pln C gene (353 bp). Bacteriocin producing lactobacilli sps can inhibit foodborne pathogens and has applications in food preservation and safety [71]. The high incidence of two non-lantibiotic bacteriocin genes (plnA and plnEF) has been reported in L. plantarum [72]. Plantaricin EF is a two-peptide bacteriocin (plnE and plnF) that depends on the complementary and synergistic action of two different peptides to function. These two-peptide bacteriocins act by binding to a specific membrane protein (a bacteriocin receptor) that leads to membrane leakage and cell death [73]. Plantaricin A is less potent antagonist than plantaricin EF, but it gives an additive antibacterial activity [74,75]. Confirmed the presence of plnEF and plnA genes in all identified L. plantarum strains. All isolates showed presence of pln EF and pln C gene.
Antioxidant activity assay by DPPH Radical Scavenging Method: DPPH radical scavenging method is widely used for the determination of antioxidant activity of the cell free supernatant of lactobacilli as its simple, rapid, sensitive and reproducible compared with other method [76]. DPPH encounters the proton-donating substances such as antioxidants, the radical is scavenged and the absorbance is reduced [77]. Strong free radical scavenging activity was observed at 18 h of incubation for most of the isolates, followed by 24 h incubation. L. fermentum L8 (74.8%) and L. plantarum L9 (72%) showed maximum activity at 18 h of incubation (Figure 11). L. fermentum L1 showed strong antioxidant activity at 6 h and L. salivarius L10 showed strong antioxidant activity at 24 h. Our findings indicated that the CFS of different strains of lactobacilli isolates produced higher antioxidant activity than control at a longer incubation time. The antioxidant compounds produced by LAB (Lactic Acid Bacteria) are reported to be acid compounds [78]. Antioxidant compounds delay, inhibit or prevent the oxidation process of fat in turn protect cells from oxidative damage by free radicals such as singlet oxygen, superoxide, peroxyl radicals, hydroxyl radical and peroxynitrite. Lactiplantibacillus plantarum produced antioxidants such as L-3- (4-Hydroxyphenyl) Lactic acid (HPLA) and L-indol-3-lactic acid [79-81]. These compounds can also contribute to antimicrobial activity) [81].
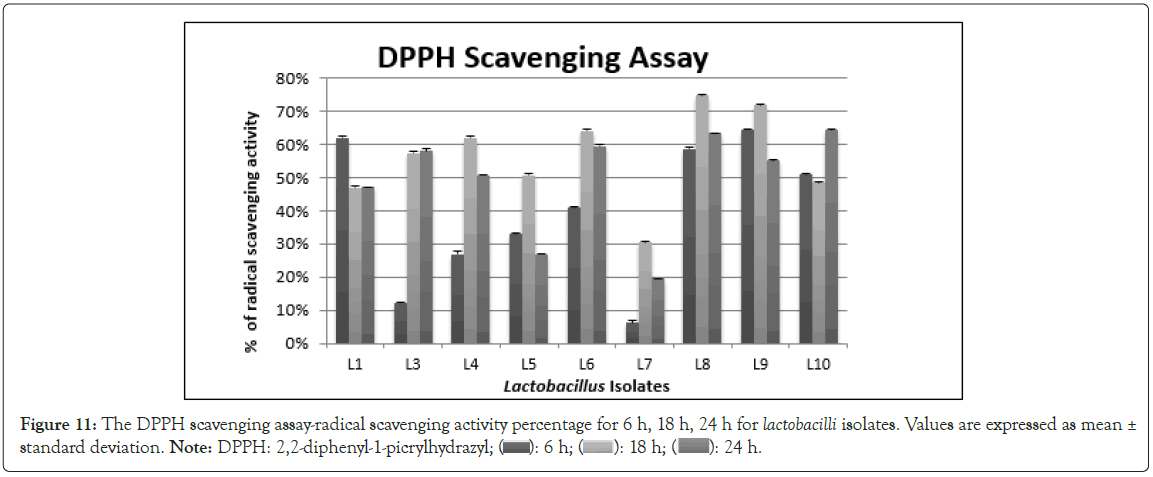
Figure 11: The DPPH scavenging assay-radical scavenging activity percentage for 6 h, 18 h, 24 h for lactobacilli isolates. Values are expressed as mean ± standard deviation. Note: DPPH: 2,2-diphenyl-1 picrylhydrazyl; ( ): 6 h; (
): 6 h; ( ): 18 h; (
): 18 h; ( ): 24 h.
): 24 h.
From our study it can be concluded that, not all human gut lactobacilli show antifungal activity. The presence of antifungal property against Aspergillus flavus MTCC 2798 was shown by ten lactobacilli strains. In vitro probiotic parameters studied showed that, lactobacilli from different individuals vary in their ability to tolerate different stress conditions, as they are exposed to different physiological circumstances. Gene study confirms the absence of virulent genes studied. Presence of bacteriocin genes plnEF and pln C was confirmed in all the isolates. Based on the invitro adhesion studies. L. fermentum MW600457 (L1), L. fermentum MW600464 (L4), L. fermentum MW600493 (L5) and L. fermentum MW485761 (L8) were considered as the potential probiotic candidates for further in vivo probiotic study. Strains with promising antifungal activity and in vitro probiotic characteristics can be used for safe food.
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
Citation: Idicula DV, George SM, Stephen JM, Parappilly SJ, Krishnaprasad R, Balan J, et al. (2024) Antifungal Human Gut Lactobacilli Isolates as Probiotic Candidate for Gut Health. J Prob Health. 12:346
Received: 23-Feb-2024, Manuscript No. JPH-24-29316; Editor assigned: 26-Feb-2024, Pre QC No. JPH-24-29316(PQ); Reviewed: 11-Mar-2024, QC No. JPH-24-29316; Revised: 18-Mar-2024, Manuscript No. JPH-24-29316(R); Published: 25-Mar-2024 , DOI: 10.35248/2329-8901.24.12.346
Copyright: © 2024 Idicula DV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which
permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.