Journal of Nutrition & Food Sciences
Open Access
ISSN: 2155-9600
ISSN: 2155-9600
Research Article - (2021)
Objective: Alpha-lactalbumin (αLA) accounts for about 20% of whey protein, which shown to have anti-inflammatory effects, but its effects on acute hepatitis have not been reported. We investigated the hepatoprotective effects of αLA in an acute hepatitis model. Since the effects of αLA on macrophages, which are related to inflammation, have not been clarified, we conducted DNA microarrays to investigate the anti-inflammatory mechanism of αLA. Methods: Sprague-Dawley rats were divided into three groups; control diet-fed group (Normal), D-galactosamine (GalN) and lipopolysaccharide (LPS)-treated and control diet-fed group (GalN/LPS), and GalN/LPS-treated, αLA- containing diet-fed group (GalN/LPS+αLA). One week after starting the test diet, rats were injected with GalN/LPS and the parameters in blood and liver were evaluated. Anti-inflammatory effect of αLA on RAW 264.7 macrophages were investigated by DNA microarray. Results: An αLA-containing diet inhibited the elevation of plasma alanine aminotransferase, aspartate aminotransferase, hepatic interleukin (IL)-6 and tumor necrosis factor-alpha (TNF-α) after the administration of GalN/LPS compared with the control diet. In RAW 264.7 cells, αLA reduced the increase of LPS-induced IL-6 and TNF-α. When the DNA microarray analysis of αLA-treated, and non-αLA-treated groups were compared, it revealed 84 genes were differentially expressed. Gene ontology analysis identified "response to LPS" and "regulation of leukocyte differentiation" as the enriched terms. Conclusion: The present study indicates that an αLA-containing diet suppresses GalN/LPS-induced hepatitis via an anti-inflammatory effect through the reduction of pro-inflammatory cytokines. It is suggested that the response of hepatic macrophages to LPS and the differentiation patterns of macrophages might be the underlying mechanism.
α-Lactalbumin; Inflammation; DNA microarray; Macrophage; Acute hepatitis
The liver is the largest organ in the human body. It is involved in metabolism, detoxification, and protein synthesis. Among the liver diseases, those that progress to hepatic cirrhosis and hepatocellular carcinoma are particularly problematic. Hepatitis B and hepatitis C account for over 90% of the cases of hepatic cirrhosis and hepatocellular carcinoma in Japan [1]. Suppression of hepatitis can help prevent the development of these diseases later on in life.
The acute liver failure model induced by administration of D-galactosamine (GalN) and lipopolysaccharide (LPS) is pathologically similar to fulminant hepatitis in humans [2]. LPS, an endotoxin, interacts with hepatic macrophages to produce tumor necrosis factor-alpha (TNF-α) and other inflammatory cytokines, causing hepatic injury [3,4]. At the same time, GalN depletes the amount of uridine nucleotides, inhibits protein synthesis in hepatocytes, and enhances the effect of LPS [5]. TNF-α, a pro- inflammatory cytokine, is the most important mediator that triggers inflammation and oxidation in hepatocytes during GalN/ LPS-induced hepatic injury [6-8].
Whey protein, which constitutes 20% of milk protein, has antioxidant, antibacterial, antiviral, anticancer, antihypertensive, and immunomodulatory properties [9-15]. It is mainly composed of β-lactoglobulin, α-lactalbumin (αLA), immunoglobulin, bovine serum albumin, and lactoferrin [16]. We previously reported that αLA, which makes up 20% of whey protein, had anti-inflammatory effect in paw exudates and adjuvant arthritis [17]. We also reported that αLA suppressed the release of pro-inflammatory cytokines after intestinal ischemia/reperfusion via the production of nitric oxide [18].
With regard to liver diseases, we reported that dietary αLA suppressed dimethylnitrosamine (DMN)-induced liver fibrosis via a nitric oxide-mediated mechanism [19]. In that study, the level of pro-inflammatory cytokines, such as TNF-α and interleukin-6 (IL-6) was not elevated, most likely because DMN-induced hepatic injury represents a chronic hepatic injury model. In contrast, GalN/LPS hepatic injury is a model of acute hepatitis, wherein TNF-α production triggers inflammation. The effect of αLA on acute hepatitis and on the inflammation caused by TNF-α, remains unclear. Therefore, in this study, we investigated the hepatoprotective effects and anti-inflammatory potential of an αLA-enriched diet in a rodent model of GalN/LPS acute hepatitis.
Macrophages are thought to be the primary effector of innate immunity and are primarily derived from monocytes. They directly phagocytose pathogens and apoptotic cells to present antigens and produce immune effectors. They are key players in the maintenance of tissue homeostasis, the formation of adaptive immune responses, inflammation, and tissue repair [20-22]. Therefore, it is considered that the evaluation of αLA in macrophages is significant in analyzing the mechanism of the inhibitory effect on hepatitis in the animal. Because the effect of αLA on macrophages, which are involved in inflammation, is not completely understood, the anti- inflammatory mechanism of αLA was examined by comprehensive analysis of gene expression in these cells using DNA microarrays.
Reagents
αLA (BioPURE Alphalactalbumin) was obtained from Davisco Foods International Inc. (Eden Prairie, MN, USA). GalN (D-galactosamine hydrochloride), LPS (from Escherichia coli O111:B4), isoflurane, Dulbecco's modified Eagle medium (DMEM), penicillin and streptomycin were obtained from Wako (Osaka, Japan). Fetal bovine serum (FBS) was sourced from Biological Industries (Cromwell, CT, USA). A commercially available diet, AIN-93 M, was purchased from Funabashi Farm Co., Ltd (Chiba, Japan); it came in two forms: AIN-93M (14% casein, Control diet) and modified AIN-93M diet (7% casein+7% αLA, αLA-containing diet) (Table 1) [23].
| Ingredients | Control diet | aLA-containing diet |
|---|---|---|
| Corn starch | 46.6% | 46.6% |
| Casein | 14.0% | 7.0% |
| a-Lactalbumin | 0.0% | 7.0% |
| a corn starch | 15.5% | 15.5% |
| Sucrose | 10.0% | 10.0% |
| Soy oil | 4.0% | 4.0% |
| Cellulose powder | 5.0% | 5.0% |
| Mineral mix (AIN-93M) | 3.5% | 3.5% |
| Vitamin mix (AIN-93) | 1.0% | 1.0% |
| L-cystine | 0.18% | 0.18% |
| Choline bitartate | 0.25% | 0.25% |
| Tertiary butylhydroquinone | 0.0008% | 0.0008% |
| Total | 100% | 100% |
Table 1: Diet compositions.
Animal experiments
Male Sprague-Dawley rats (6-weeks-old; weighing 160-180 g) were obtained from SLC Japan, Inc. (Shizuoka, Japan) and were used in the following experiments after 1 week of acclimation in a room maintained at 22 ± 1°C with a humidity of 55 ± 15% and a 12-h light/dark cycle, with the light period running from 7:00 to 19:00. All animal experiments were approved by the Meiji Co., Ltd. Institutional Animal Care and Use Committee (Tokyo, Japan) and were performed in accordance with the guidelines of Meiji Co., Ltd. for the care and use of laboratory animals (approval code: 2017_3871_0078, 14 June 2017).
The rats were randomly divided into three groups: (1) control diet- fed group (normal); (2) GalN/LPS-treated and control diet-fed group (GalN/LPS); (3) GalN/LPS-treated, αLA-containing diet-fed group (GalN/LPS+αLA). One week after starting the diets, rats were intraperitoneally injected with GalN/LPS (400 mg GalN+30 μg LPS/kg body weight), whereas rats in the normal group were injected with saline. Blood samples were obtained from the saphenous vein at 2, 4, 6, and 8 h after the GalN/LPS injection. The rats were euthanized under isoflurane anesthesia at 10 h after the GalN/LPS injection. At euthanasia, blood samples were collected from the abdominal aorta and centrifuged for 15 min at 1,500 × g. Plasma was then collected and the liver was excised, and kept frozen at -80°C for subsequent biochemical analysis.
Biochemical analysis of plasma and liver tissue
The activities of plasma aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured by colorimetric slides using Fuji Dri-Chem NX500 (Fujifilm, Tokyo, Japan). Samples of the medial lobe of the liver were homogenized with a three-fold volume of potassium phosphate buffer, and then centrifuged at 10,000 × g for 10 min at 4°C. The resulting supernatants were used to measure the concentrations of IL-6 and TNF-α. Hepatic IL-6 and TNF-α levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Minneapolis, MN, USA) following the manufacturer’s protocol. The protein content of the supernatant was determined by BCA Protein Assay Kit (Takara, Tokyo, Japan).
Cell culture
Murine macrophage cell line, RAW264.7, was obtained from ATCC (Manassas, VA, USA) and cultured at 37°C in DMEM supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified 5% CO2 atmosphere.
Cell viability
RAW264.7 cells were seeded in a 96-well plate at a density of 5 × 104 cells/well, and cultured for 24 h. After treatment with αLA (100, 500, 1000 μg/mL) for 24 h, the viability of cells was determined by the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer's instructions.
Measurement of cytokine levels
RAW264.7 cells were seeded in a 96-well plate at a density of 5 × 104 cells/well, and cultured for 24 h. After treatment with αLA (100, 500, 1000 μg/mL) for 24 h and subsequent stimulation with 20 ng/mL LPS for an additional 6 h, the amounts of TNF-α and IL-6 in the culture supernatants were determined using ELISA kits (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's instructions.
Microarray analysis
RAW264.7 cells were seeded in a 12-well plate at a density of 5 × 105 cells/well, and cultured for 24 h. After treatment with αLA (1000 μg/mL) for 24 h, RNA was isolated from RAW264.7 cells using a Maxwell RSC simplyRNA Cells kit (Promega, Madison, WI, USA) and analyzed on a Maxwell RSC Instrument according to the manufacturer's instructions. The quantity and quality of extracted RNA were evaluated by measuring the absorbance at 260:280 nm using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The pure extracted RNAs with 260:280 ratios >1.8 were used for microarray analysis. Each sample was additionally run on the Agilent Bioanalyzer RNA Nano 6000 chip to further assess the quality and integrity of the total extracted RNA. All samples had an RNA integrity number (RIN) >7. cDNA was synthesized from RNA using the GeneChip Whole Transcript (WT) Amplification kit (Thermo Fisher Scientific, Waltham, MA, USA) as described by the manufacturer. The sense cDNA was then fragmented and biotin-labeled with terminal deoxynucleotidyl transferase (TdT) using the GeneChip WT Terminal labeling kit. The samples were then subjected to Affymetrix microarray analysis with an Affyetrix Clariom™ S Assay, Mouse. Approximately 5.5 μg of labeled DNA target was hybridized to the Affymetrix GeneChip Array at 45°C for 16 h. The hybridized arrays were washed and stained on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner (Affymetrix). Array data export processing and analysis was performed using Affymetrix® GeneChip Command Console® Software (AGCC). The software was Affymetrix Power Tools (affymetrix-power-tools.html) and R 3.5.1 (https://www.r-project.org/).
The data were summarized and normalized with Signal Space Transformation-Robust Multichip Analysis (SST-RMA) method implemented in Affymetrix® Power Tools (APT). We exported the results with gene level SST-RMA analysis and performed analysis for determining the DEGs.
Statistical significance of the expression data was determined using independent t-test and fold change in which the null hypothesis was that no difference exists among the groups. False discovery rate (FDR) was controlled by adjusting the p value using the Benjamini- Hochberg algorithm. For a DEG set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene enrichment and functional annotation analyses for significant probe list was performed using Gene Ontology (http://geneontology.org). The GO terms were chosen if (1) their adjusted p-value (adj. p-value) was less than 0.05, and (2) term_size was between 10 to 500. All data analyses and visualization of DEGs were conducted using R.
Statistical analysis
Data are expressed as means ± standard error of the mean (SEM). The homogeneity of variance was tested by Bartlett's test. When variances were homogeneous, the data were analyzed by one-way analysis of variance (ANOVA) and followed by a Tukey-Kramer test. When variances were not homogeneous, the data were analyzed by Kruskal-Wallis test and followed by a Steel-Dwass test. A p<0.05 was considered statistically significant.
Since AST and ALT are the most commonly used markers of liver damage, these were measured. The effect of αLA on GalN/ LPS-induced increase in the activities of plasma AST and ALT is presented in Figure 1. The activities of these enzymes at 4, 6, 8, and 10 h after GalN/LPS injection were significantly higher in the GalN/LPS group than in the normal group. At the same time points, the activities in the GalN/LPS+αLA group were significantly lower than those in the GalN/LPS group.
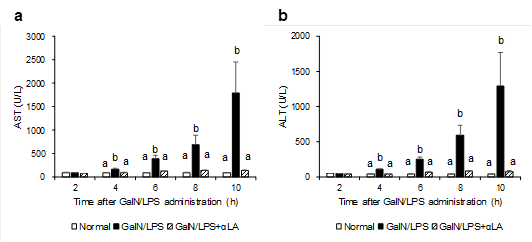
Figure 1: Effects of aLA on the plasma levels of AST (a), ALT (b) in GalN/LPS-treated rats. Rats were fed a standard AIN-93M diet or an aLA-containing diet for 7 days. After one week from starting the diets, rats were intraperitoneally injected with GalN/LPS or saline. Blood samples were obtained at 2, 4, 6, 8, and 10 h after the GalN/LPS injection. The activities of AST and ALT in plasma were measured by colorimetric slides using Fuji Dri-Chem NX500. Each value represents the mean ± SEM (n=4-8). The different letters at each measured time represent the statistical differences at p<0.05 among the groups by Tukey-Kramer test or Steel-Dwass test.
When GalN/LPS hepatotoxicity develops, TNFα is first elevated, followed by IL-6, and when hepatocytes are damaged, ALT and AST are elevated. Therefore, we measured inflammatory cytokines associated with elevated ALT and AST, which are markers of liver injury. The effect of αLA on the GalN/LPS-induced increase in hepatic TNF-α and IL-6 at 10 h after GalN/LPS injection is presented in Figure 2. The GalN/LPS-induced increase in hepatic TNF-α level was attenuated by pretreatment with αLA (Figure 2a). The levels of IL-6 were significantly lower in the GalN/LPS+αLA group than in either the normal or GalN/LPS group (Figure 2b).
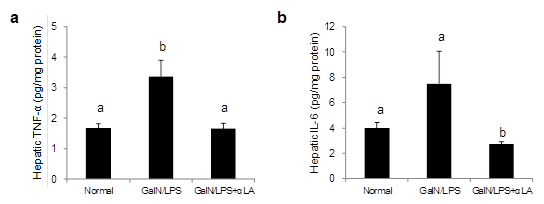
Figure 2: Effects of aLA on the hepatic levels of TNF-a (a) and IL-6 (b) in GalN/LPS-treated rats. Rats were fed a standard AIN-93M diet or an aLA-containing diet for 7 days. After one week from starting the diets, rats were intraperitoneally injected with GalN/LPS or saline. Livers were excised at 10 h after the GalN/LPS injection. Hepatic TNF-a and IL-6 were measured by ELISA. Each value represents the mean ± SEM (n=4-8). The different letters at each measured time represent the statistical differences at p<0.05 among the groups by Tukey-Kramer test or Steel-Dwass test.
Since inflammatory cytokines produced by hepatic macrophages are key factors in inflammation and act during liver inflammation, the inflammatory cytokines TNF-α and IL-6 were measured as indicators of inflammation in the in vitro study. In the absence of LPS treatment, αLA did not exhibit any cytotoxic effect on RAW264.7 macrophages up to a concentration of 1000 μg/mL (data not shown). Pretreatment with αLA significantly inhibited the production of TNF-α and IL-6 induced by LPS in a dose-dependent manner (Figure 3). These results indicate that αLA inhibited the LPS-induced production of inflammatory cytokines in RAW 264.7 macrophages at non-toxic concentrations.
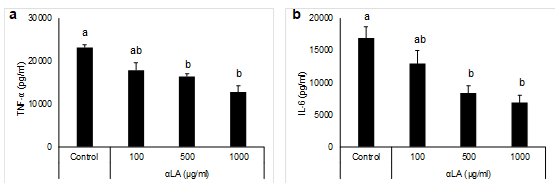
Figure 3: Effects of aLA on LPS-stimulated production of TNF-a (a) and IL-6 (b) in RAW 264.7 macrophages. Cells were preincubated with 100-1000 µg/mL aLA for 24 h and then treated with 20 ng/mL LPS for 6 h. Concentrations of TNF-a and IL-6 in the cultured medium were determined by ELISA. Each value represents the mean ± SEM of three independent experiments. The different letters at each measured time represent the statistical differences at p<0.05 among the groups by Tukey-Kramer test or Steel-Dwass test.
Microarray analysis of RAW 264.7 cells treated with 1,000 μg/ mL of αLA for 24 h revealed different patterns of expression upon hierarchical clustering (Figure 4). A total of 84 differentially expressed genes (DEGs) were identified (fold change ≥ 1.5, ≤-1.5, p<0.05). Of these, 62 genes were upregulated and 22 were downregulated by treatment with αLA. The top 20 genes whose expression was enhanced or suppressed are shown in Tables 2 and 3.
| Gene symbol | Fold-change | p-value | Gene description |
|---|---|---|---|
| Myc | 3.1761 | 4.5.E-04 | myelocytomatosis oncogene |
| Olfml3 | 2.6369 | 9.6.E-04 | olfactomedin-like 3 |
| Abca1 | 2.564 | 6.5.E-04 | ATP-binding cassette, sub-family A (ABC1), member 1 |
| Itgax | 2.5163 | 3.5.E-02 | integrin alpha X |
| Egr2 | 2.4663 | 2.4.E-03 | early growth response 2 |
| Egr1 | 2.3076 | 2.2.E-03 | early growth response 1 |
| Dcstamp | 2.2408 | 2.8.E-02 | dentrocyte expressed seven transmembrane protein |
| Mmp12 | 2.2008 | 6.8.E-04 | matrix metallopeptidase 12 |
| Atp6v0d2 | 2.0057 | 4.6.E-05 | ATPase, H+ transporting, lysosomal V0 subunit D2 |
| Il13ra2 | 1.9762 | 1.2.E-03 | interleukin 13 receptor, alpha 2 |
| Nes | 1.9701 | 7.9.E-04 | nestin |
| Ocstamp | 1.9532 | 7.7.E-03 | osteoclast stimulatory transmembrane protein |
| Prss46 | 1.9484 | 2.3.E-04 | protease, serine 46 |
| Tmem26 | 1.9303 | 4.8.E-03 | transmembrane protein 26 |
| Il1rn | 1.9064 | 2.5.E-05 | interleukin 1 receptor antagonist |
| Irg1 | 1.8817 | 4.0.E-04 | immunoresponsive gene 1 |
| Cd3d | 1.7601 | 1.3.E-02 | CD3 antigen, delta polypeptide |
| Plk2 | 1.7526 | 9.0.E-03 | polo-like kinase 2 |
| Olfr1045 | 1.7348 | 3.1.E-03 | olfactory receptor 1045 |
| Dusp1 | 1.7178 | 2.3.E-03 | dual specificity phosphatase 1 |
Table 2: List of the 20 genes upregulated by aLA.
| Gene symbol | Fold-change | p-value | Gene description |
|---|---|---|---|
| Tsga13 | -1.9957 | 4.7.E-02 | testis specific gene A13 |
| Angptl3 | -1.8219 | 1.6.E-02 | angiopoietin-like 3 |
| Gm10377 | -1.7667 | 1.7.E-02 | predicted gene 10,377 (Gm10377), mRNA. |
| Sspn | -1.7232 | 2.7.E-02 | sarcospan |
| Cldn34-ps | -1.7211 | 4.0.E-04 | claudin 34, pseudogene |
| Plekhg3 | -1.6966 | 1.0.E-02 | pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| Tbx22 | -1.6404 | 3.6.E-02 | T-box 22 |
| Abcd2 | -1.6278 | 6.0.E-03 | ATP-binding cassette, sub-family D (ALD), member 2 |
| Gm19549 | -1.6143 | 2.7.E-02 | PREDICTED: predicted gene, 19,549 (Gm19549), miscRNA. |
| Vmn1r72 | -1.6143 | 4.1.E-02 | vomeronasal 1 receptor 72 |
| Gas6 | -1.6081 | 1.6.E-03 | growth arrest specific 6 |
| Ift122 | -1.6048 | 7.0.E-03 | intraflagellar transport 122 |
| Dgkg | -1.5796 | 3.7.E-02 | diacylglycerol kinase, gamma |
| Rangrf | -1.5602 | 1.3.E-02 | RAN guanine nucleotide release factor |
| Nccrp1 | -1.5396 | 1.3.E-02 | non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| Gpr22 | -1.5382 | 4.3.E-02 | G protein-coupled receptor 22 |
| Tlr8 | -1.5359 | 4.1.E-02 | toll-like receptor 8 |
| Vmn2r71 | -1.5286 | 1.7.E-02 | vomeronasal 2, receptor 71 |
| Tgtp2 | -1.5215 | 3.8.E-02 | T cell specific GTPase 2 |
| Tll1 | -1.5187 | 2.1.E-04 | tolloid-like |
Table 3: List of the 20 genes downregulated by aLA.
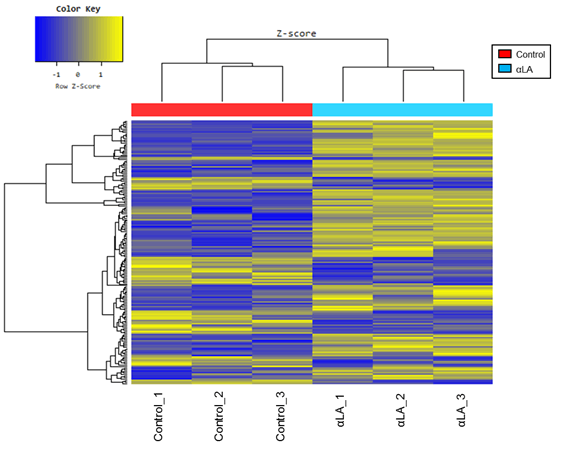
Figure 4: Heat map of differentially expressed genes (DGEs) with fold-change expression >1.5. RAW 264.7 macrophages were treated with 1000 µg/mL aLA for 24 h (n=3) or with the vehicle control (n=3). Rows represent genes and columns represent samples. Yellow blocks represent high and blue blocks represent low expression relative to the vehicle-treated cells.
Gene ontology is a method of expressing gene function. It expresses the function of individual genes by combining several terms [24]. After treatment with αLA, the top 14 GO terms involved in changes in gene expression is shown in Table 4. The GO terms with a greater number of altered genes were “response to lipopolysaccharide,” “response to molecule of bacterial origin,” “cellular response to biotic stimulus,” “cellular response to lipopolysaccharide,” “cellular response to molecule of bacterial origin,” and “regulation of leukocyte differentiation.”
| Gene ontology term | Term | No.genes1 | Adjusted p-value |
|---|---|---|---|
| response to lipopolysaccharide | GO:0032496 | 9 | 0.032 |
| response to molecule of bacterial origin | GO:0002237 | 9 | 0.036 |
| cellular response to biotic stimulus | GO:0071216 | 7 | 0.036 |
| cellular response to lipopolysaccharide | GO:0071222 | 7 | 0.036 |
| cellular response to molecule of bacterial origin | GO:0071219 | 7 | 0.036 |
| regulation of leukocyte differentiation | GO:1902105 | 7 | 0.039 |
| response to glucocorticoid | GO:0051384 | 6 | 0.036 |
| response to corticosteroid | GO:0031960 | 6 | 0.038 |
| response to retinoic acid | GO:0032526 | 5 | 0.036 |
| anion homeostasis | GO:0055081 | 4 | 0.036 |
| phospholipid homeostasis | GO:0055091 | 3 | 0.021 |
| negative regulation of cell division | GO:0051782 | 3 | 0.032 |
| membrane disruption in other organism | GO:0051673 | 3 | 0.032 |
| natural killer cell differentiation | GO:0001779 | 3 | 0.039 |
Note: The number of transcripts overexpressed in each GO category. |
|||
Table 4: Enrichment of gene GO categories.
GalN and LPS are often used to generate a model of acute hepatitis in rodents [8,25]. Such a model is useful for assessing the hepatoprotective function of various food ingredients [26-29]. GalN/LPS-induced liver injury involves the immune response and is characterized by the infiltration of mononuclear cells in the liver, as well as increased activities of plasma AST and ALT [8,30,31]. Indeed, in this study, the activities of ALT and AST were significantly elevated following the administration of GalN/LPS, but the increase was suppressed in the GalN/LPS+αLA group.
TNF-α is the most important inflammatory cytokine produced during GalN/LPS-induced hepatic injury [32,33]. It triggers the inflammatory cascade that induces other cytokines, such as IL-6 and nitric oxide [6,34]. Various agents have been shown to prevent GalN/LPS-induced hepatic injury by regulating the activity of TNF-α [35,36]. Here, hepatic TNF-α and IL-6 were elevated following the administration of GalN/LPS; however, the increase was suppressed by pretreatment with αLA. We previously reported that αLA has an anti-inflammatory effect, manifested as decreased release of cytokines in rodent models of ischemia-reperfusion, as well as in colon inflammation and carcinogenesis induced by azoxymethane and dextran sodium sulfate [17,37]. Our present findings are in agreement with previous results in that αLA showed an anti-inflammatory effect by suppressing TNF-α and IL-6.
It was shown in a previous study that intact αLA is present in plasma after its administration into the duodenum [18]. This study indicated that oral αLA might have been absorbed in the intact form and had a direct influence on the liver. Also, amino acids and peptides released after the digestion and absorption of αLA might have contributed to the effects that were observed by us. αLA contains a large number of aspartic acid/asparagine, cysteine, glycine, histidine, isoleucine, leucine, lysine, threonine, and tryptophan residues compared to casein. Some of these amino acids have been reported to have a hepatoprotective effect. For example, a mixture of cysteine, methionine, and serine improved the hepatotoxicity induced by acetaminophen in mice [38]. Moreover, branched-chain amino acids have been shown to have beneficial effects on hepatic ischemia-reperfusion-induced hepatic injury in rats [39]. Some dietary amino acids, such as asparagine, glutamine, glycine, histidine, lysine, serine, and tryptophan have hepatoprotective effects on GalN-induced hepatitis [40]. It has been reported that lysine, tryptophan, histidine, and arginine inhibited the expression of inflammatory mediators by suppressing NF-κB activity in cultured hepatocytes and GalN/LPS-injected rats [41]. Imbalances in the levels of amino acids in plasma result in a specific profile of amino acid concentrations, which is a useful diagnostic or predictive marker for particular pathological conditions [42]. Therefore, to prevent pathological conditions, an appropriate amino acid supply could be effective [43]. Amino acids derived from the αLA-containing diet used in the present study might play a role in maintaining plasma amino acid equilibrium, and exert a hepatoprotective effect against acute hepatic injury.
αLA-containing diet significantly inhibited the LPS/GalNinduced elevation of plasma ALT, AST, and hepatic TNF-α and IL-6 in rats. These results suggest that αLA inhibits the production of inflammatory cytokines and suppresses hepatic injury. Macrophages are cells that are primarily responsible for the production of cytokines. Therefore, we investigated the antiinflammatory mechanism of αLA using mouse macrophage, RAW 264.7 cells. αLA significantly reduced the increase in the production of TNF-α and IL-6 by LPS. These results are consistent with those of animal studies. DNA microarray was subsequently carried out to comprehensively analyze the effect of αLA on the expression of genes in macrophages.
To our knowledge, this is the first study to investigate the effects of αLA on macrophages using comprehensive gene expression analysis. Comparative analysis was carried out using αLA-treated and untreated groups. For the DNA microarray data, we first performed a hierarchical cluster analysis and found that the gene expression patterns differed between the αLA-treated and untreated groups. The following genes with altered expression levels are described below: myelocytoma tumor gene (Myc), ATP-binding cassette transporter A1 (Abca1), ATPase, H+ transport, lysosomal V0 subunit D2 (Atp6v0d2), and Toll-like receptor 8 (Tlr8).
Myc was upregulated by αLA treatment. Myc functions as a transcription factor that promotes the expression of numerous target genes to regulate cell death, proliferation, and metabolism [44,45]. Myc induces the expression of a major subset of genes associated with alternative activation programs in concert with IL-4 downstream signaling mediators, signal transducer and activator of transcription-6 (STAT6), and peroxisome proliferator-activated receptor γ (PPARγ) [46]. These studies indicate that Myc may exert immunomodulatory functions in macrophages. It is possible that the increased expression of Myc in this study also activated the expression of various genes and showed adaptability of the immune response to LPS administration after αLA treatment.
Abca1 was upregulated by αLA treatment. LPS-stimulated inflammatory responses are enhanced in Abca1-deficient macrophages and Abca1-deficient mice [47-50]. In addition, increased Abca1 expression has been reported to enhance the efflux of LPS [51]. It is possible that increased Abca1 expression was involved in reducing the inflammatory response to LPS and in promoting the excretion of LPS in the present study.
The expression of Egr2 was enhanced by αLA treatment. Stimulation of M2 macrophage polarization by IL-4 and IL-13 results in an increase in the expression of Egr2 [52]. M2 macrophages express anti-inflammatory cytokines, such as IL-10, and are involved in the regulation of inflammation and post-inflammatory tissue repair [53,54]. It has also been reported that EGR2, together with EGR3, induces the expression of anti-inflammatory regulators, such as SOCS1 and SOCS3 [55,56]. Because the expression of Egr2 was increased in the present study, it is possible that αLA treatment increased the polarization in M2 macrophages and controlled inflammation.
Atp6v0d2 was also upregulated by αLA treatment. ATP6V0D2 plays a central role in the acidification of intracellular vesicles [57]. In ATP6V0D2-deficient cells, LPS, a TLR4 agonist, increased the production of inflammatory cytokines and NF-κB activation [58]. These results indicate that ATP6V0D2-dependent intravesicular acidification is required for internalization of TLR4 and may be important in preventing excessive LPS-induced inflammation. ATP6V0D2 has also been reported to promote autophagosomelysosomal membrane fusion and may be important for clearance of bacteria [59].
The expression of Tlr8 was decreased by αLA treatment. TLR8 recognizes single-stranded RNA from viruses and bacteria [60]. The recognition of TLR8 acts, via Myd 88, on the downstream mitogenactivated protein kinase (MAPKs) and IκB kinase (IKK) complexes to increase the expression of NF-κB [61]. Increased expression of NF-κB promotes the production of inflammatory cytokines, such as TNFα and IL-6. Algal extracts containing polyunsaturated fatty acids and dyes have also been reported to reduce inflammation in LPS-stimulated THP-1 macrophages, with reduced expression of TLR8 [62]. In the present study, it is possible that the decreased expression of Tlr8 by treatment with αLA reduced the binding of Tlr8 ligand to Tlr8, resulting in the suppression of inflammation.
GO analysis is a standard method in many high-throughput experimental studies. In this study, "response to LPS" and "regulation of leukocyte differentiation" was detected as the enriched GO terms. The genes for Myc, Abca1, Atp6v0d2, and Tlr8, whose expression is increased or decreased as described above, are involved in the function of the detected GO terms.
Microarray analysis of αLA-treated macrophages revealed changes in the expression of various genes involved in LPS response and leukocyte differentiation. We believe that the mechanism by which dietary administration of αLA suppressed GalN/LPS-induced liver injury in animal studies may be related to changes in the responsiveness of liver macrophages to LPS and the differentiation potential of macrophages.
The purpose of this study was to investigate the anti-inflammatory effect of an αLA-containing diet on galactosamine and lipopolysaccharide-induced acute hepatitis. Furthermore, we conducted DNA microarray analysis of the effects of αLA on macrophages to comprehensively analyze gene expression and to investigate the mechanism of anti-inflammatory effects of αLA.
In conclusion, the present study indicates that an αLA-containing diet suppresses GalN/LPS-induced hepatitis via an anti- inflammatory effect through the suppression of the elevation of pro-inflammatory cytokines, such as TNF-α and IL-6. In addition, it is suggested that the response of hepatic macrophages to LPS and the differentiation patterns of macrophages might be the underlying mechanism. αLA may be a new therapeutic candidate for the suppression of acute liver injury.
This research was financially supported by Meiji Co., Ltd.
Akika Fukawa, Orie Kobayashi and Makoto Yamaguchi are employees of Meiji Co. Ltd. Akira Hosono has no conflicts of interest directly relevant to the content of this article.
Citation: Fukawa A, Kobayashi O, Yamaguchi M, Hosono A (2021) Anti-inflammatory Effect of Alpha-Lactalbumin on Lipopolysaccharide-stimulated RAW 264.7 Macrophages and Galactosamine/Lipopolysaccharide-Induced Liver Injury in Rats. J Nutr Food Sci. 11:807.
Received: 14-Jun-2021 Accepted: 28-Jun-2021 Published: 05-Jul-2021
Copyright: © 2021 Fukawa A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : This research was financially supported by Meiji Co., Ltd.