Poultry, Fisheries & Wildlife Sciences
Open Access
ISSN: 2375-446X
ISSN: 2375-446X
Research Article - (2019)Volume 7, Issue 2
The Cypriniformes comprises of 5 families (Catostomidae, suckers; Cyprinidae, minnows; algae eaters; Gyrinocheilidae, Balitoridae, river loaches Cobitidae, loaches), depending on investigators every one with subfamilies poorly separate (e.g., 426 generas and 2 to 12 subfamilies recognized Substantial amounts of amino acid and DNA data involved with mtDNA can be provided for phylogenetic analyses [1]. Previous researcher resolved the phylogenetic relationship of Schizothoracinae fishes only for the one species comparison with the other 21 species [2]. Increasing the number of mitochondrial genome sequences of Schizothoracinae and reconstructing the Schizothoracinae phylogenetic tree based on a more comprehensive dataset are necessary to address the remaining problematic clades So analysis of Phylogenetics of Cyprinid taxa based on the functionally vital genes can help to understand the functional divergence and speciation [2]. Majority of the whole mitochondrial genomic sequence occupied by 13 proteincoding genes. In the present study, we our recent study based on monophyletic origin of the Schizothoracinae fish in respect to mitochondrial protein-coding genes, to find out the phylogenetic relationship of Schizothoracinae fishes from the available genome upto date on the basis of mitochondrial protein coding genes.
Present study was conducted in Panjkora River, it is reflected as the chief life line of lower Dir, and it is Malakand division part and situated in the province of Khyber Pakhtunkhwa Pakistan. It lies in the range of Hindu Kush region between Latitude: 34° 39' 59.99" N, Longitude: 71° 45' 59.99" E. Fish samples of Schizothorax plagiostomus were collected from River panjkora using different types of nets namely hand nets, cast nets and hooks. Transferred the specimen to the lab of department of zoology were the samples were stored. For the amplification of the mtDNA of Schizothorax plagiostomus we used standard high salt extraction method, we extracted the mtDNA from the preserved muscle tissues in 95% ethanol [3]. For polymerase chain reaction (PCR) amplification sixteen sets of primers were designed which were based on original mitochondrial genomes DNA sequences of cyprinid fish. Cocktail reaction included The 25 μL to 6 μL of 10 × buffer, every nucleotide of 1.5 μL (dNTP), every primer of 1 μL, Taq DNA polymerase about 1.5 unit, template DNA of 1–2 μL. The process of thermocycling was started for 5 min at 94°C, followed through 20 cycles at 94°C for 30 s, 56°C for 50 s, and 72°C for 1 min 30 s, with 0.1°C reducing the temperature for annealing to every cycle, with the annealing temperature at 54°C Then 12 other cycles used to, there was an end of final cycle at 8 min extension. On 1.2% of Agarose gel in 1×Trisacetate-EDTA buffer for all samples, PCR product was electrophoresed about 1 μl at 80 V for 30 min then staining with ethidium bromide, as well as in the Gel-Doc system visualized under Ultra violet illumination. The purified PCR products through standard protocols then for sequencing sent to the Sangon biotech company (Sangon Biotech Company Shanghi).
Analysis of sequence
Using the program Clustal W the DNA sequences were aligned [4]. The DNA sequences was edited and analysed with Auto Assembler (Applied Bio systems) and DNASIS (Hitachi Software Engineering Co. Ltd) [5]. The locations of the 13 protein-codding genes were determined by comparisons of the amino acid sequences. The phylogenetic association was inferred by MEGA 6.0 [6]. For the phylogenetic association, the mtDNA, 13 sequenced protein coding genes of Schizothorax plagiostomus were used, and the sequences of other cyprinids were used for infringe the phylogenetic relationships there sequences retrieved from NCBI.
Phylogenetic analyses
In addition to the newly obtained sequence of S. plagiostomus, we obtained sequences of 13 protein coding genes for another 31 species of 8 genera in Schizothoracinae to elucidate among members of the subfamily Schizothoracinae the phylogenetic relationships. Sequences of 13 protein coding to serve as out-groups genes of four species in the subfamily Barbinae and three species in Cryprininae were obtained from GenBank in the subsequent phylogenetic analyses. In Table 1 all species and the respective GenBank numbers used in the present study are listed. All 13 protein coding genes of all species the alignments were combined, and a concatenated alignment was then generated. Using ClustalW thirteen protein-coding gene sequences were translated into their corresponding amino acids and with default settings aligned, and translated into the nucleotide sequence for obtaining the better results. Gabs and missing data determined and was deleted finally we left with datasets the concatenated nucleotide sequences of the 13 protein-coding genes, were generated for the subsequent phylogenetic analyses. Phylogenies were reconstructed using maximum-likelihood tree constructed by using the mega software. Following accession numbers were taken. (KT184924), (KT210882.1), (NC_022866.1), (NC_023366.1), (NC_027670.1), (KP892531.1),(NC_026294.1)(NC_024621.1),(NC_017873.1),( NC_031537.1),(NC_026205.1),(KT223584.1) (NC_023829.1), (NC_020781.1), (KC513574.1), (JX202592), (NC_020339.1), (NC_029708.1), (NC_027162.1), (KJ577589.1), (NC_021448.1), (NC_031803.1), (NC_024833.1), (KT944287.1), (NC_020360.1), (KR527479), (KM593242), (KC558499), (KC351895), (JX232379), (KP316067), (KF597526), (KM268050), (KR527479), (KF528985), (KF976395), (KC558497), (JQ004279), (KC558498), (AB239595), (JQ004278), (KM659026), (KF564793), (KJ476998), (GU170401), (AP009047), (AB238965), (AB239600)(JX311437), (NC_026294.1).
| Family | Subfamily | Genus | Species | GenBank no |
|---|---|---|---|---|
| Ingroup | Schizothoracinae | Schizothorax | Schizothorax esocinus | KT210882.1 |
| Cyprinidae | Schizopyge niger | NC_022866.1 | ||
| Schizothorax progastus | NC_023366.1 | |||
| Schizothorax kozlovi | NC_027670.1 | |||
| Schizothorax yunnanensis | KP892531.1 | |||
| Schizothorax lantsangensis | NC_026294.1 | |||
| Schizothorax chongi | NC_024621.1 | |||
| Schizothorax biddulphi | NC_017873.1 | |||
| Schizothorax nepalensis | NC_031537.1 | |||
| Schizothorax davidi | NC_026205.1 | |||
| Schizothorax nukiangensis | KT223584.1 | |||
| Schizothorax prenanti | NC_023829.1 | |||
| Schizothorax oconnori | NC_020781.1 | |||
| Schizothorax waltoni | KC513574.1 | |||
| Schizothorax waltoni 2 | JX202592 | |||
| Schizothorax macropogon | NC_020339.1 | |||
| Schizothorax graham | NC_029708.1 | |||
| Schizothorax lissolabiatus | NC_027162.1 | |||
| Schizothorax dolichonema | KJ577589.1 | |||
| Schizothorax richardsonii | NC_021448.1 | |||
| Schizopyge gongshanensis | NC_031803.1 | |||
| Schizothorax pseudoaksaiensis | NC_024833.1 | |||
| Schizothorax labiatus | KT944287.1 | |||
| Schizothorax wangchiachii | NC_020360.1 | |||
| Schizopygopsis | Schizopygopsis malacanthus | KR527479 | ||
| Schizopygopsis malacanthus baoxingensis | KM593242 | |||
| Schizopygopsis thermalis | KC558499 | |||
| Schizopygopsis younghusbandi | KC351895 | |||
| Schizopygopsis younghusbandi 2 | JX232379 | |||
| Schizopygopsis pylzovi | KP316067 | |||
| Ptychobarbus | Ptychobarbus dipogon | KF597526 | ||
| Ptychobarbus kaznakovi | KM268050 | |||
| Schizopygopsis malacanthus | KR527479 | |||
| Oxygymnocypris | Oxygymnocypris stewartii | KF528985 | ||
| Gymnodiptychus | Gymnodiptychus pachycheilus | KF976395 | ||
| Gymnocypris | Gymnocypris dobula | KC558497 | ||
| Gymnocypris eckloni | JQ004279 | |||
| Gymnocypris namensis | KC558498 | |||
| Gymnocypris przewalskii | AB239595 | |||
| Gymnocypris przewalskii ganzihonensis | JQ004278 | |||
| Diptychus | Diptychus maculatus | KM659026 | ||
| Aspiorhynchus | Aspiorhynchus laticeps | KF564793 | ||
| Outgroup | ||||
| Cyprinidae | Cyprininae | Carassius | Carassius auratus | KJ476998 |
| Carassius gibelio | GU170401 | |||
| Cyprinus | Cyprinus carpio | AP009047 | ||
| Barbinae | Barbus | Barbus barbus | AB238965 | |
| Barbus trimaculatus | AB239600 | |||
| Puntius | Puntius chalakkudiensis | JX311437 |
Table 1: List of species and sequences used in this study.
Combined data set of all the 13 protein-coding genes of 31 species of 8 genera in Schizothoracinae species of Schizothorax yielded two clades having out groups and groups species showed closest relationships with each other. Schizothorax lantsangensis group showed 99% of the posterior nodal probability with the Schizothorax waltoni group and also showed the maximum probability with Schizothorax chongi group and Schizothorax lantsangensis group. Maximum bootstrap values supports our results showed in Figure 1.
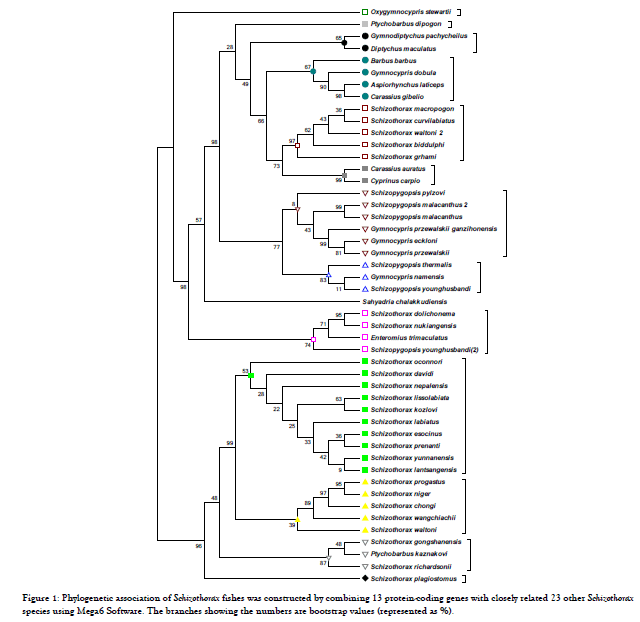
Figure 1. Phylogenetic association of Schizothorax fishes was constructed by combining 13 protein-coding genes with closely related 23 other Schizothorax species using Mega6 Software. The branches showing the numbers are bootstrap values (represented as %).
Current study was conducted to find out the phylogenetic relationship of Schizothoracinae fishes. The S. plagiostomus mitochondrial genome sequence was found to be 16569 bp length. Like other cyprinids fishes, genome of S. plagiostomus consisted of 13 protein coding gene (PCGs), 22 TRNAs gene, two ribosomal RNA (12S rRNAs and 16S rRNAs) genes, one control region and light strand replication origin (OL) consisted of 33 bp. The neighbour joining analysis was performed in MEGA6 with 1000 bootstrap replicates [7,8]. Consequently, in inter-and intraspecific phylogeny in an animal’s mitochondrial genome the genetic information offered, in the studies it is widely analysed [9]. Phylogenetic relationships among genera and species under Schizothoracinae have been investigated based on morphological characters, RAPD analysis. Cyprinid fishes phylogenetic analysis based on the functionally vital genes can support to know the functional divergence and speciation of these fishes Consequently, correspondingly it is the set of proteins which have the greatest marked variances between species observed, Monophyly is intensely supported for the of schizothorax fishes in the set of protein [10,11]. Schizothoracinae fishes showed the closest relationship with the other fishes formed a different cluster. It was found that the species belonging to the northern Himalayas grouped together while species from north-eastern Himalayas remained separate [12]. Maximum bootstrap values supports our results, these results are somewhat similar with the finding earlier [8]. Inferred the phylogenetic relationship on the basis of whole mitochondrial data set.
As the recent developments in molecular techniques based on genes are very much useful for establishing taxonomical and phylogenetic relationships among different species. The present study resolved the phylogenetic relationships among the 48 species of subfamily Schizothoracinae, study of phylogenetic relationship on the basis of sequences of protein-coding gene in these species offers useful visions to the phylogenetic status of cyprinid fishes and provide The step for further investigations of issues with taxonomic and conservation and phylogenetic in this vital up of fishes.
The authors alone are responsible for the content and writing of the paper. The authors report no conflicts of interest.
Citation: Bibi S, Khan MF, Rehman A, Nouroz F (2019) Polyphyletic Origins of Schizothoracinae Fishes (Cyprinidae) in Respect to Their Mitochondrial Protein-Coding Genes. Poult Fish Wildl Sci 7:207. doi: 10.35248/2375-446X.19.7.207
Received: 03-Sep-2019 Accepted: 24-Oct-2019 Published: 31-Oct-2019 , DOI: 10.35248/2375-446X.19.7.207
Copyright: © 2019 Bibi S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.