Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research - (2022)Volume 11, Issue 6
Hollyhock is one important ornamental plant grown in garden beds across the world. It is susceptible to many diseases caused by diverse pathogens. Among these viral pathogens are causing enormous damage to the hollyhock. The aim of the present study was to identify the begomovirus and DNA satellites associated with the Yellow Vein Mosaic (YVM) and Enation Leaf Curl (EnLC) disease complex of hollyhock. Sample from hollyhock plants exhibiting a typical begomovirus-like symptoms were collected from Pusa campus, New Delhi (India). To know the status of the begomovirus associated with the infected hollyhock samples, total genomic DNA isolated was subjected to PCR amplification using virus specific primer pair. The partial genome (1.2 kb) sequencing of begomovirus associated with ten hollyhock samples (five each from YVM and EnLC pheotype) indicates they shared more homology with begomovirus, Cotton leaf curl Multan virus (CLCuMuV). Nucleotide identities between ten hollyhock samples are more than 95 percent. Therefore, two representative samples (H1and H2) showing YVM and one showing EnLC (H3) were selected for full-length genome amplification of the virus using RCA method. The samples showed no amplificatiobn for the DNA-B specific primers and gave amplification to betasatellite ans alphasatellite specific primers, indicating the begomovirus associated with hollyhock as monopartite begomovirus. The pairwise comparision of complete genome of the three begomovirus using Sequence Demarcation Tool (SDT) showed highest nucleotide (nt) identity of 88.0 to 92.7 percent of their genome with CLCuMuV, 92.5-96 per cent of betasatellites sequences with Ludwigia Leaf Distortion Betasatellite (LuLDB) and 90.4 to 93.2 percent of alphasatellites sequences with Ageratum Enation alphasatellite (AEV). Further recombinantion analysis showed that the begomovirus and DNA satellites under study are recombinants originated from previously reported begomoviruses and DNA-satellites.
Hollyhock; Begomoviruses; DNA-sattelites; Recombinantion; Phylogentic analysis; Sequence demarcation tool; Polymerase chain reaction
Genus, Begomovirusismost important among the fourteen genera divided based on genome structure, replication, vector transmission and host range in the family, Geminiviridae [1]. Members of the genus, Begomovirus are the major group of plant viruses affecting different crops worldwide. These are transmitted by complex morphological indistinguishable cryptic species of whiteflies [2] in a persistent circulative and non-propagative manner. Further, based on the genome structure of the begomoviruses were classified into into Old World (OW) and New World (NW) begomoviruses. Most of begomoviruses reported so far belongs to the OW begomovises having single genome component (monopartite) and remaining belongs to NW begomoviruses having two genome components (bipartite) [3]. However, recent past several bipartite begmoviruses are reported from the OW as well [4]. The OW begomoviruses are more diverse compared NW and they have AV2 gene, which is absent in NW begomoviruses [3]. The bipartite begomoviruses encodes seven to eight proteins from their genome (DNA-A and DNA-B). Whereas, monopartite begomoviruses encode five to six proteins from its genome, aslo referred as homolougue of DNA-A of bipartite viruses. In bipartite begomoviruses, genes coding for CP and Rep, TrP and REn are present on DNA-A component and genes for movement of the viral particle (MP and NSP) are present on DNA-B component [5].
Most of the monopartite begomoviruses are additionally associated with classes of circular ssDNA satellites (beta, alphasatellite and delta) with their genome [4,6,7]. Eventhough, most of bipartite begomoviruses reported earlier are shown their genome to be not associated with satellites, off late ther are reports supporting the evidence of association DNA satellite molecules with their genomes [8]. The beta and deltasatellites (ssDNA) are true satellites; since they are depend on the helper virus for multiplication, movement and vector transmission [9]. Whereas alphasatellites are not ture satellite, because they are autonomous in replication, however depend on the helper virus for movement and encapsidation [10].
Hollyhock (Alcea rosea L., family, Malvaceae) is one of the important ornamental plant grown in garden beds throughout the world. Ornamental hollyhock plant was known to be originated from Asia and Europe. Hollyhock plants are having very broad leaves and grown to increase the aesthetic value of the gardens. Apart from its aesthetic value, its flowers are used in the treatment of chest complaint and decoction is used to improve blood circulation [11]. Further, hollyhock leaves and flowers are also used for preventing and treating breathing disorders and digestive tract problems. Despite its many uses, the ornamental plant is a natural host for many viruses [12-18] which causes severe damage to hollyhock. Therefore, the current study was attempted to characterize begomovirus associated with the Yellow Vein Mosaic (YVM) and Enation Leaf Curl (EnLC) disease of hollyhock in India.
Virus source plants and vector transmission
During the suvery five samples each from hollyhock plants exhibiting yellow vein mosaic and enation leaf curl symptoms were collected from Pusa campus, New Delhi, India during the year 2018 and 2019 (Figures 1a-1c). Two asymptomatic samples from the same location were also collected. During sample collection, presence of huge number of whiteflies on undersurface of the leaves hollyhock plants was observed. These plants were showing typical begomovirus like-symptoms indicating that hollyhock plants might have been infected with begomovirus. A part of leaf sample was used for transmission of disease by using whiteflies (Bemisia tabaci) reared on healthy cotton plants under controlled conditions as described by Venkataravanappa et al. [19]. Adult whiteflies were allowed separately to feed on the infected hollyhock plant leaf samples showing typical enation leaf curl and yellow vein mosaic disease collected during the survey with an acquisition access period of 24 h. Then the whiteflies (ten numbers on each) were transferred to ten days old seedlings of hollyhock (ten) with an inoculation access period of 24 h in separate cages. After this inoculated plants were sprayed with insecticide (0.05% imidocloprid) and maintained separately in insect-proof net house for symptom development. The transmission was repeated multiple times by using newly inoculated plants after symptoms expression as source plant. The pure culture of the yellow vein mosaic and enation leaf curl was maintained under insect-proof net house for futher use. The presence of begomovirus in the samples collected from the hollyhock plants showing yellow vein mosaic and enation leaf curl after incolucation using whiteflies from the field sample was further confirmed through PCR using degenarative primer pairs specific to begomovirus.

Figure 1: Holly hock plants showing (a) mild yellow mosaic (b) complete yellow vein mosaic, (b) leaf curl symptoms under natural conditions.
DNA extraction, polymerase chain reaction, and sequence analysis
To assess the status of suspected begomovirus causing YVM and EnLC disease in hollyhock, total genomic DNA was extracted from five each hollyhock samples with EnLC and YVM along with two asymptomatic hollyhock leaf samples by CTAB method [20]. The extracted DNA from the sample was subjected to the PCR assay using the begomovirus genome specific primers specific to DNA-A/Homolouge of DNA-A [21] and DNA-B [22] with the expected amplicons of 1.2 kb and 1.5 kb, respectively. Complete genome of begomovirus from the two samples with YVM (H1,H2) and one sample with EnLC (H3) symptoms was amplified by RCA (rolling circle amplification) as decribed by Haible et al. [23]. In order to obtain monomeric circular genome of the virus showing YVM and EnLC disease symptoms, 2 µl of RCA product was digested with BamH1 and purified using Gel extraction kit. The purified RCA products of YVM and EnLC samples of hollyhock were cloned into linearised plasmid (pUC19) [19] and transformed to Escherichia coli DH 5 α cells. Two clones from each sample were confirmed by restriction digestion and colony PCR using virus specific primers. Further, complete genome of DNA-satellites (alpha and betasatellites) associated with infected hollyhock plants was amplified by PCR using universal primer pairs specific to DNA-satellites [9,24]. The PCR amplified DNA-satellites products were cloned into pTZ57R ⁄T vector (Thermo Fisher Scientific), transformed to Escherichia coli DH 5 α cells and confirmed for the presence of insert by restriction digestion and colony PCR using virus specific primers. Selected clones were sequenced in both orientations from the commercial sequencing service provider.
Analysis of viral genome sequences and recombination breakpoint events
Sequence similarity check for the obtained begomoviruses, beta and alphasatellites sequences was done at NCBI using BLASTn. Sequences of the viruses showed maximum blast score with present hollyhock isolates and other closely related begomoviruses were retrieved from the NCBI GenBank (all viruses’ acronyms used in this study as per the Walker et al.). The pair wise identity score between hollyhock infecting begomoviruses and satellites with selected begomoviruses and DNA-satellites was calculated by using SDT version 1.2 [25]. Phylogenetic tree was drawn using the maximum likelihood method with 1,000 bootstrapped replications available in MEGA 7 software [26]. Recombination events detection in the viral genome and DNA satellites was carried out using RDP4 with default settings [27].
Disease survey and detection of begomovirus and associated DNA-satellites
Hoolyhock plants exhibiting symptoms of YVM and EnLC were observed at PUSA Campus, NewDelhi. The symptoms produded were mutually exclusive with plant producing symptoms of YVM doesn’t have EnLC symptoms and vice-versa. Five sample from each symptom phenotype were collected separately during survey was confirmed with begomovirus infection by PCR using specific primers with expected amplicon of 1.2 kb in all the ten symptomatic samples and no amplification observed in the asymptomatic samples.
Attempt to amplify DNA-B didn’t resulted in the amplificon expected; indicating the begomovirus associated with hollyhock samples is monopartite. The partial genome sequence analysis of begomovirus obtained from 1.2 kb amplicons from ten infected hollyhock samples (5 EnLC and 5 YVM) showed the association of monopartite DNA virus species (nt identity >97% among the EnLC and YVM samples). Further, identity of DNA-A sequences of virus from hollyhock was 100 between them. Therefore, three representative samples YVM (H1, H2 from YVM samples and H3 from EnLC) were selected to amplify the complete genome of the begomovirus (2.7 kb) by RCA method.
PCR amplification for DNA-satellite molecules from the three selected hollyhock samples (H1, H2 and H3) infected assocated begomoviruses with primers specific to beta and alphsatellites [9,24] resulted in amplicons of 1.2 to 1.3 kb in size specific, respectively from the three samples. This indicated the indicating the association of DNA satellites with YVM and EnLC disease of hollyhock plants.
Whitefly, B.tabci transmission of begomovirus associated with hollyhock plants showing YVM and EnLC
Separte whitefly, B.tabaci transmission experiments for samples of hollyhock showing EnLC and YVM to healthy holloyhock was successful (10/10 plants) in each case. Inoculated plants developed similar symptoms at 20-25 days post-inoculation (dpi). Control experiment using asymptomatic samples did not showed any symptoms on inoculated healty hollyhock plants.
Sequencing
The complete genome sequences of begomovirus associated with hollyhock was obtained from cloned RCA products from three samples of (H1 and H2 with YVM symptoms and H3 with EnLC). BLAST analyses revealed 99-100 percent nucleotide identity of DNA-A of the virus among three clones considered for sequencing form H1, H2 and H3 samples. Similarly, 1.3 kb and 1.2 kb in size PCR products specific to complete genome of betasatellites (H1β, H2β, and H3β) and alphasatellites (H1D1, H2D1 and H3D1) associated with the begomovirus obtained from three hollyhock samples were cloned and sequenced.
The sequence data (DNA-A and DNA-satellites) obtained from the three hollyhock samples (H1, H2, H3) were assembled by using different omics programs (Sea View, Bioedit and Clustal X2). The consensus sequences of DNA-A (Acc. no. MN127817-19), alphsatellites (Acc No. MN127823-25) and betsatellites (Acc. No. MN127820-22) were submitted NCBI Genbank.
Genome organization of begomovirus associated with YVM and EnLC of hollyhock
The complete viral genomes of three hollyhock isolates (H1, H2, H3) were determined to be 2738 to 2750 nts long and genome structure of the hollyhock isolates (H1, H2, H3) is analogous to other monopartite DNA virus from OW with potential encode five conserved ORFs: two ORFs: (V2 and V1), on plus strand and four ORFs (C1, C2, C3, C4) present on minus strand of genome component of the virus. The plus strand and minus strand of homologous DNA-A was separated by an Common Region (CR), which contained highly conserved nonanucleotide (TAATATTAC), sequence nicked by the rolling-circle initiator protein (Rep protein) to initiate replication. The Rep protein (encoded by C1) of the hollyhock isolates have all conserved domains similar other DNA viruses as described by Vadivukarasi et al. [28], except the GRS motif (RFFDLISPTRSAHFHPNIQRAKS).
The pairwise comparison of complete genome of the three hollyhock isolates (H1, H2, H3) with other selected begomoviruses retrieved from the NCBI database using SDT revealed that, these isolates shared maximum nt identity of 88.0 to 92.7% to CLCuMuV (JN678803) infecting cotton in India and <88% nt identity with several other begomoviruses infecting different crops in India. This result is also supported by pairwise identity scores calculated by SDT between hollyhock isolates and other begomoviruses available NCBI database (Figures 2a-2c). Based on genome sequence comparison and guidelines given by ICTV Study group [1] , the three hollyhock isolates (H1, H2, H3) are distinct strains of Cotton leaf curl Multan virus (CLCuMuV) for which the additional descriptor [India: New Delhi: Hollyhock: 2018] is proposed.
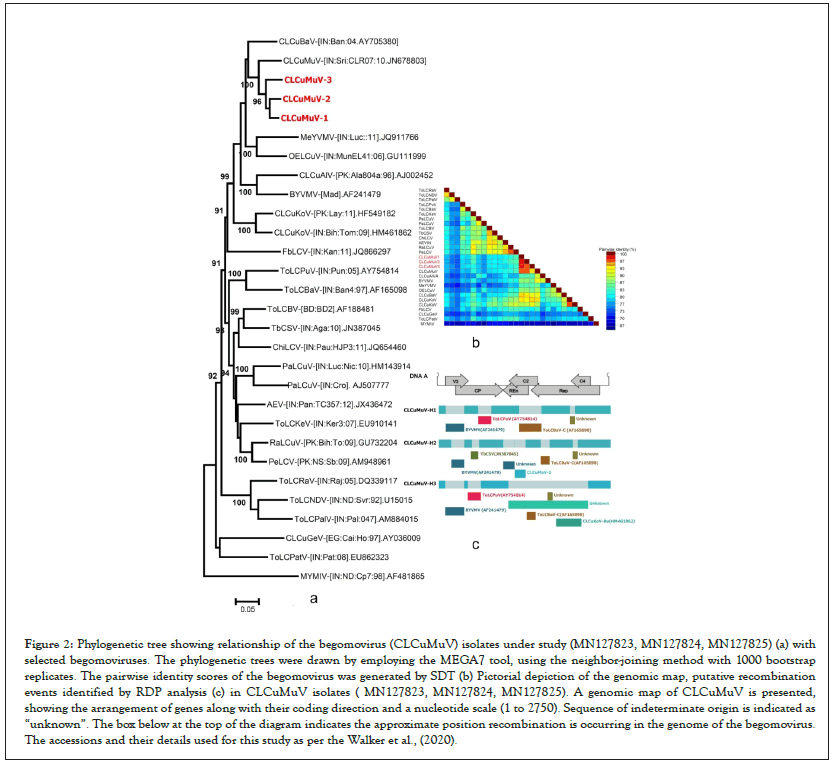
Figure 2: Phylogenetic tree showing relationship of the begomovirus (CLCuMuV) isolates under study (MN127823, MN127824, MN127825) (a) with selected begomoviruses. The phylogenetic trees were drawn by employing the MEGA7 tool, using the neighbor-joining method with 1000 bootstrap replicates. The pairwise identity scores of the begomovirus was generated by SDT (b) Pictorial depiction of the genomic map, putative recombination events identified by RDP analysis (c) in CLCuMuV isolates ( MN127823, MN127824, MN127825). A genomic map of CLCuMuV is presented, showing the arrangement of genes along with their coding direction and a nucleotide scale (1 to 2750). Sequence of indeterminate origin is indicated as “unknown”. The box below at the top of the diagram indicates the approximate position recombination is occurring in the genome of the begomovirus. The accessions and their details used for this study as per the Walker et al., (2020).
Phylogenetic analysis was carriedout by comparing complete genome of the three isolates (H1, H2, H3) obtained in the present study with 29 known begomoviruses infecting different crops available in NCBI database. Result showed that three hollyhock isolates (H1, H2, H3) isolates in the same clade along with CLCuMuV (JN678803) infecting cotton in India (Figure 2a). Similarly, ORFs V1 and C2 of three hollyhock isolates (H1, H2, H3) shared the clade with CLCuMuV (JN678803), ORF C1 shared the clade with CLCuBaV (AY705380) and CLCuKoV (HF549182) and ORF C4 shared with CLCuKoV (HF549182) clade (data not shown). However, ORFs V1 shared a clade with BYVMV (AF241479) (data not shown). The analyses clearly showed that genome of three isolates (H1, H2, H3) associated hollyhock is probably arose as a result of recombination with other cotton infecting begomoviruses.
Genome organization of betasatellite associated with YVM and EnLC of hollyhock
The length of the three betasatellites (H1β, H2β and H3β) (under accession number MN127820, MN127821 and MN127822) ranged from 1363 to 1371bp. Betasatellites sequences from hollyhock showed typical features of other betasatellites reported on different crops [2,29] . Sequence analysis of three betasatellites characterized in the study showed nt identity of 92.5-96.7% with LuLDB isolates originating from the Indian subcontinent infecting okra and hibiscus (Figure 3a). Based on the proposed criteria for species demarcation threshold for betasatellites, set at nt identity 91% [1], three betasatellites (H1β, H2β, and H3β) identified in hollyhock are an isolates of LuLDB. This result was also supported by SDT showing three betasatellites closely related to LuLDB infecting different crops.
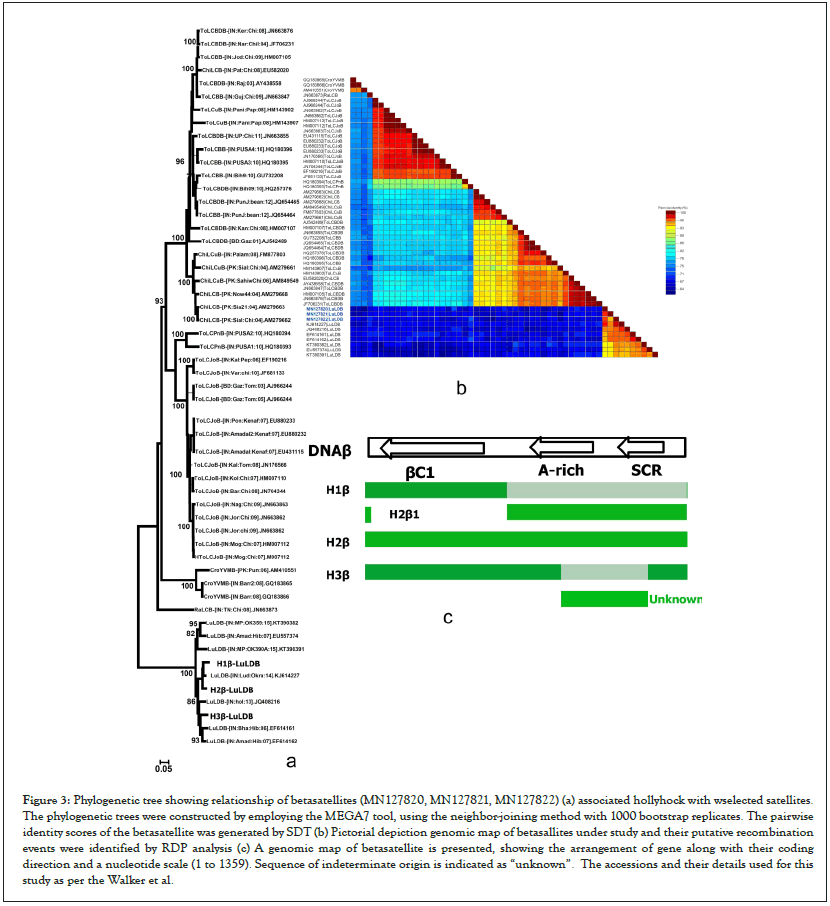
Figure 3: Phylogenetic tree showing relationship of betasatellites (MN127820, MN127821, MN127822) (a) associated hollyhock with wselected satellites. The phylogenetic trees were constructed by employing the MEGA7 tool, using the neighbor-joining method with 1000 bootstrap replicates. The pairwise identity scores of the betasatellite was generated by SDT (b) Pictorial depiction genomic map of betasallites under study and their putative recombination events were identified by RDP analysis (c) A genomic map of betasatellite is presented, showing the arrangement of gene along with their coding direction and a nucleotide scale (1 to 1359). Sequence of indeterminate origin is indicated as “unknown”. The accessions and their details used for this study as per the Walker et al.
Phylogenetic analysis of complete nucleotide sequences of three betasatellites (H1β, H2β, H3β) isolated from hollyhock with selected betasatellites revaled that three betasatellites (H1β, H2β, H3β) closely clustered with previously reported LuLDB isolates originating from the Indian subcontinent for which a full-length sequences are available in the NCBI database (Figures 3a-3c).
Genome organization of alphasatellite associated with YVM and EnLC of hollyhock
Sequence analysis of three alphasatellites (H1D1, H2D1, H3D1) (under accession number MN127823, MN127824, MN127825) isolated from hollyhock determined their length to be 1364 to 1371 bp in length. Alphasatellites in the current study have similar characterstic features of other alphasatellites reported so far on different crops [30]. Sequence analysis revealed that two alphasatellites (H1D1 and H2D1) shared maximum nt identity of 90.4 to 93.2% with AEA (HG518790, FR772085) infecting okra and hollyhock, where as one alphasatellite (H3D1) is showed more homology with AEA (HE599396) infecting cotton (Figures 4a-4c). Based on proposed criteria for species demarcation threshold for alphasatellite [31], the alphasatellites identified here are an isolates of AEA infecting okra, hollyhock and cotton and belongs genus, Colecusatelliteof the family, Alphasatellitidae. This results was also well supported by the phylogenetic analysis showing two alphasatellites (H1D1, H2D1) clustered with several isolates of AEA (HG518790 and FR772085) infecting okra and hollyhock, where as one alphasatellite (H3D1) is closely clustered with AEA (HE599396) infecting cotton. This result was also supported by SDT, in which three alphasatellite isolated from hollyhock plants are closely related to AEA (Figure 4a).
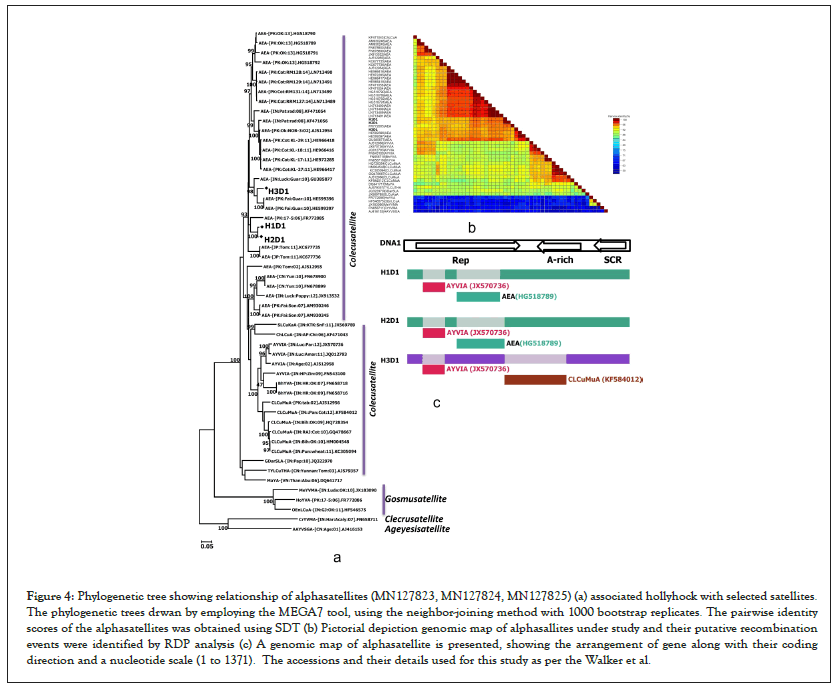
Figure 4: Phylogenetic tree showing relationship of alphasatellites (MN127823, MN127824, MN127825) (a) associated hollyhock with selected satellites. The phylogenetic trees drwan by employing the MEGA7 tool, using the neighbor-joining method with 1000 bootstrap replicates. The pairwise identity scores of the alphasatellites was obtained using SDT (b) Pictorial depiction genomic map of alphasallites under study and their putative recombination events were identified by RDP analysis (c) A genomic map of alphasatellite is presented, showing the arrangement of gene along with their coding direction and a nucleotide scale (1 to 1371). The accessions and their details used for this study as per the Walker et al.
Recombination
For recombination break point analysis analysis of three hollyhock isolates (H1, H2 and H3) understudy, sequences of CLCuMuV, CLCuKoV, CLCuAIV, CLCuBaV, PaLCuV, ToLCBaV, TbCSV and BYVMV isolates were used (Table 1). RDP4 with default settings was used to identify recombination break points [27]. RDP analysis showed the evidence of intra and inter specific recombination in three hollyhock isolates (H1, H2, H3) with most of the coding and non-coding regions of its genome derived from different begomoviruses viz., CLCuMuV, CLCuKoV, CLCuAIV, CLCuBaV, PaLCuV, ToLCBaV, TbCSV and BYVMV as major and minor parents (Figure 2a). Similarly, RDP analysis was done for betasatellites (H1β, H2β and H3β) genome and it revealed evidence of intra and inter-specific recombination suggesting most of betasatellites DNA descended from LuLDB infecting okra and hibiscus crops (Figure 3a). Further, RDP analysis for alphasatellites (H1D1, H2D1 and H3D1) indicates the exchange of gene fragment from SLCuKaA, AYVIA and AEA as donor parents infecting okra, hollyhock and cotton (Figure 4a). All viruses and satellites acronyms used in this study are as per the Walker et al.).
| DNA-A | Break point begin-end a | Parent-like sequences | P-Values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Major Parent | Minor parent | RDP | GENECOV | Max Chi | Chimera | Si Scan | 3Seq | |||||||
| CLCuMuV-H1 | 132-482 | CLCuBaV-[IN:Ban:04].AY705380 | BYVMV[Mad].AF241479 | 3.224 × 10-6 | 7.975 × 10-6 | 2.533 × 10-6 | 7.38 × 10-6 | - | 3.573 × 10-6 | |||||
| 547-745 | CLCuMuV-Dar[IN:Sri:CLR07:10].JN678803 | ToLCPuV-[IN:Pun:05].AY754814 | 7.055 × 10-4 | - | - | - | 1.973 × 10-2 | 4.969 × 10-5 | ||||||
| 1304-1733 | PaLCuV-Cro[IN:Cro].AJ507777 | ToLCBaV-C[IN:Ban4:97].AF165098 | 3.436 × 10-2 | - | 8.31 × 10-1 | - | 2.481 × 10-6 | 2.197 × 10-2 | ||||||
| 2509-2753 | CLCuAlV-Al[PK:Ala804a:96].AJ002452 | ToLCBV-[BD:BD2].AF188481 | 6.916 × 10-3 | - | - | 2.312 × 10-2 | - | 9.37 × 10-4 | ||||||
| CLCuMuV-H2 | 235-482 | CLCuBaV-[IN:Ban:04].AY705380 | BYVMV[Mad].AF241479 | 3.224 × 10-6 | 7.975 × 10-6 | 2.533 × 10-6 | 7.38 × 10-6 | - | 3.573 × 10-6 | |||||
| 1315-1468 | CLCuMuV-H1 | CLCuMuV-H2 | - | - | 1.829 × 10-5 | - | 3.808 × 10-4 | 1.732 × 10-6 | ||||||
| 1647-1673 | PaLCuV-Cro[IN:Cro].AJ507777 | ToLCBaV-C[IN:Ban4:97].AF165098 | 3.436 × 10-2 | - | 8.31 × 10-1 | - | 2.481 × 10-6 | 2.197 × 10-2 | ||||||
| 2059-2156 | CLCuAlV-Al[PK:Ala804a:96].AJ002452 | ToLCBV-[BD:BD2].AF188481 | 6.916 × 10-3 | - | - | 2.312 × 10-2 | - | 9.37 × 10-4 | ||||||
| CLCuMuV-H3 | 121-482 | CLCuBaV-[IN:Ban:04].AY705380 | BYVMV[Mad].AF241479 | 3.224 × 10-6 | 7.975 × 10-6 | 2.533 × 10-6 | 7.38 × 10-6 | - | 3.573 × 10-6 | |||||
| 1315-2737 | CLCuMuV-H1 | CLCuMuV- [IN:Sri:CLR07:10].JN678803 | 1.406 × 10-4 | 1.938 × 10-2 | 1.242 × 10-22 | 1.479 × 10-26 | 2.336 × 10-10 | 5.277 × 10-67 | ||||||
| 1658-1835 | PaLCuV-Cro[IN:Cro].AJ507777 | ToLCBaV-C[IN:Ban4:97].AF165098 | 3.436 × 10-2 | - | 8.31 × 10-1 | - | 2.481 × 10-6 | 2.197 × 10-2 | ||||||
| 2059-2156 | CLCuAlV-Al[PK:Ala804a:96].AJ002452 | ToLCBV-[BD:BD2].AF188481 | 6.916 × 10-3 | - | - | 2.312 × 10-2 | - | 9.37 × 10-4 | ||||||
| 2339-2826 | CLCuMuV-H2 | CLCuKoV-[IN:Bih:Tom:09].HM461862 | 5.38 × 10-15 | 3.104 × 10-4 | 3.395 × 10-10 | - | - | 5.524 × 10-23 | ||||||
| DNAD1(Alphasatellite) | ||||||||||||||
| H1D1 | 132-326 | SLCuKaA-[IN:KTK:SnF:11].JX569789 | AYVIA-[IN:Luc:Par:12].J × 570736 | 4.298 × 10-3 | 3.346 × 10-2 | 9.407 × 10-4 | 2.955 × 10-3 | 8.286 × 10-5 | 1.047 × 10-2 | |||||
| 419-792 | AEA-[IN:Pat:rad:08].KF471054 | AEA-[PK:OK:13].HG518789 | 3.122 × 10-4 | 2.173 × 10-2 | 5.601 × 10-3 | 4.94 × 10-3 | 2.684 × 10-4 | 9.061 × 10-5 | ||||||
| H1D2 | 133-326 | SLCuKaA-[IN:KTK:SnF:11].JX569789 | AYVIA-[IN:Luc:Par:12].J × 570736 | 4.298 × 10-3 | 3.346 × 10-2 | 9.407 × 10-4 | 2.955 × 10-3 | 8.286 × 10-5 | 1.047 × 10-2 | |||||
| 418-830 | AEA-[IN:Pat:rad:08].KF471054 | AEA-[PK:OK:13].HG518789 | 3.122 × 10-4 | 2.173 × 10-2 | 5.601 × 10-3 | 4.94 × 10-3 | 2.684 × 10-4 | 9.061 × 10-5 | ||||||
| H1D3 | 128-321 | SLCuKaA-[IN:KTK:SnF:11].JX569789 | AYVIA-[IN:Luc:Par:12].J × 570736 | 4.298 × 10-3 | 3.346 × 10-2 | 9.407 × 10-4 | 2.955 × 10-3 | 8.286 × 10-5 | 1.047 × 10-2 | |||||
| 856-39 | AEA-[PK:OK:13].HG518792 | CLCuMuA-[IN::Pan:Cot:12].KF584012 | 6.894 × 10-10 | 6.298 × 10-5 | 5.741 × 10-5 | 5.653 × 10-6 | 7.936 × 10-6 | 2.54 × 10-6 | ||||||
| DNAβ (Betasatellite) | ||||||||||||||
| H1β | 856-39 | H3β | H2β | 6.99 × 10-14 | 2.68 × 10-17 | 2.577 × 10-17 | 8.719 × 10-15 | 3.494 × 10-21 | 7.566 × 10-32 | |||||
| H2β (Non-recombinant) | ||||||||||||||
| H3β | 958-1138 | LuLDB-[IN:Lud:Okra:14].KJ614227 | LuLDB-[IN:hol:13].JQ408216 | 2.936 × 10-5 | 9.25 × 10-7 | 4.33 × 10-4 | 8.195 × 10-9 | 2.866 × 10-6 | 1.717 × 10-10 | |||||
Note: NS-Recombination Non-significance; a -The text in the parenthesis of this column indicates ORF’s in which break points are identified.
Table 1: Details of recombination between CLCuMuV and DNA satellites associated with hollyhock and other begomoviruses detected using RDP4.
Hollyhock is an important winter ornamental plant grown in different garden beds to increase the asthetic value throughout India. The plant is not only had ornamental value, it also a good source of medicine. In the present study, samples from hollyhock plants showing the YVM and EnLC symptoms are confirmed with begomovirus association by PCR assay. Complete viral genome sequence analysis indcated the diseased hollyhock plants associated with CLCuMuV infecting cotton in the Indian subcontinent. CLCuMuV was first identifed in 1967 in the Multan district of Pakistan on scattered cotton (Gossypium hirsutum) plants [32, 33]. Form their it was spread rapidly to other cotton-growing regions of Pakistan and Indian subcontinent. The virus (CLCuMuV) not only infecting cotton, it also infects other important horticutlural crops [34-36]. All begomoviruses associated with cotton leaf curl disease dentified from India are infecting cotton, okra, and tomato. Infection of cotton viruses to malvaceous and non-malvaceous hosts may be due to high inoculum load of cotton viruses and its vector whitefly cryptic species. Hollyhock is malvaceous plant, which is commonly grown as ornamental plant in the garden as well as pots in front of houses. Due to existing morphologically indistinguishable whitefly cryptic species complex in India, the cotton infecting virus may easily spreading to other crops including hollyhock. Similarly, the litrature showed the diverse viruses infecting ornamental plants worldwide [37,38]. But information regarding begomovirus infections on ornamental plants is very scanty from India [39]. This might be due to the neglected study of the ornamental crops viruses while carrying out surveys.
Betasatellites and alphastellies are found associated with both OW mono and bipartite DNA viruses in many crops [10, 40-42]. In the present the hollyhock samples are shown to be associated withLuLDB, which is also identified in begomoviruses infecting different crops in India [17,43-44] . Phylogenetic analysis showed that betasatellite associated hollyhock close relationship with LuLDB infecting begomoviruses in solanaceous crops. This clearly indicates the possible spread of the betasatellite across geographic region. The study also showed that betasatellites does not contain iteron sequences, therefore still it depend on helper virus for its replication [45-47].
The alphasatellites (H1D1, H2D1, H3D1) identified in present study are closely related to the AEA infecting okra, hollyhock and cotton [48,49]. Which are having similar features of other alphastellites identified in many crops [29,41,42]. Alphasatellites play major role in attenuation of disease symptoms and are known to be involved in the maintenance of low level of betasatellites accumulation in the plant [50]. Further, they were also implicated in suppression of RNAi pathway in begomovirus disease complexes [4]. In the study the hollyhock plants at New Delhi exhibiting YVM and EnLC symptoms are mutually exclusive, which might be due the difference in the associated DNA satellites with plants. This phenomena was well documented in plants infected with begomoviruses and DNA satellites in combination [4,29,41,42].
Recombination is one of the key factor for evolution and creating genetic diversity in begomoviruses [51]. The results showed that recombination has led to the formation of a distinct strain of CLCuMuV and its satellites infecting hollyhock in India. The recombination analysis suggested that CLCuMuV isolates have obtained at least some of its sequences from the previously reported begomoviruses infecting different crops. The overall the results of the recombination and phylogenetic analysis suggested that CLCuMuV and its satellites are evolved from different monopartite DNA viruses (CLCuMuV, CLCuKoV, CLCuAIV, CLCuBaV, PaLCuV, ToLCBaV, TbCSV, BYVMV), betastellites (LuLDB), and alphasatellies (SLCuKaA, AYVIA and AEA) reported previously in India on different crops (all viruses acronyms used in this study as per the Walker et al.).
The present study clearly indicating that the CLCuMuV is expanding its host range by infecting hollyhock to other malvaceous hosts and becoming a serious threat for cultivation of many ornamental and horticulture crop plants. Hence there is a need for more comprehensive survey to identify possible spread of the begomovirus infections in the country to assess role of ornamental plants in serving as virus reservoirs for the crop plant infecting virus diseases. This will form the basis of our future investigations.
The research was supported by project on “Consortium platform on Vaccines and diagnostics”, Indian Council of Agricultural Research, Government of India, New Delhi, India.
VV and KVA conducted the research. CNLR, VV and KSS prepared and edited the manuscript, SH, HDV and NM did the bioinformatics analysis and MKR provided the overall guidance of the work and provided the facility.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Ashwathappa KV, Venkataravanappa V, Nandan M, Hiremath S, Vinaykumar HD, Shankarappa KS, et al. (2022) Association of Begomovirus and DNA-Satellites Complex with Enation Leaf Curl and Yellow Vein Mosaic Disease of Hollyhock. Virol Mycol. 11:245.
Received: 03-Oct-2022, Manuscript No. VMID-22-19452 ; Editor assigned: 06-Oct-2022, Pre QC No. VMID-22-19452 (PQ); Reviewed: 20-Oct-2022, QC No. VMID-22-19452 ; Revised: 27-Oct-2022, Manuscript No. VMID-22-19452 (R); Published: 03-Nov-2022 , DOI: 10.35248/2161-0517.22.11.245
Copyright: © 2022 Ashwathappa KV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.