Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2023)Volume 16, Issue 4
Background: Host-microbiome dysbiosis have been linked to Type 2 Diabetes Mellitus (T2DM). The purpose of this paper is to investigate whether Phascolarctobacterium and Phascolarctobacterium faecium (P. faecium) serve as ideal biomarkers for T2DM. On this basis, to evaluate the key role of multi-omics analysis in the early diagnosis of T2DM.
Method: Detected stool samples from healthy people, T2DM patients and T2DM patients after metformin treatment in our cohort study by 16S ribosomal Ribo-Nucleic Acid (rRNA) gene amplicon sequencing. In addition, various baseline clinical and metabolic index were collected to evaluate the diagnostic models of Receiver Operator Characteristic (ROC) curves which combined use of intestinal bacteria, fatty acids and micro Ribo-Nucleic Acid (miRNA) as predictive tools for early detection of T2DM.
Results: Our multi-omics analysis indicates that T2DM patients had specific gut microbiota dysbiosis, where faecium and P. faecium are correlated with multiple biochemical indicators of T2DM and the intervention of metformin had some influence on the composition of gut microbiota. We also identified the diagnostic models of ROC curves were able to classify T2DM patients from healthy people with a better estimation accuracy.
Conclusion: Phascolarctobacterium and P. faecium can be novel biomarkers for the early diagnosis of T2DM. The multi-omic analysis based on gut microbiome provides insights for elucidating the specific mechanism in the host-microbiome dysbiosis at the early metabolic disorders.
Multi-omics analysis; Phascolarctobacterium; Phascolarctobacterium faecium; ROC curves; Type 2 diabetes mellitus.
Type 2 Diabetes Mellitus (T2DM) is one of the fastest growing global health emergencies in the 21st century. The pathogenesis of T2DM involves metabolomics, transcriptomics, microbiology and other almost all life activities [1,2]. Prevention and early diagnosis are the most effective health strategies at present. Composition and metabolic stability of intestinal micro ecological and specific miRNAs expression in human are both essential for early development of T2DM has been repeatedly reported in the last years [3,4]. A large number of studies approved the pathogenic effects of gut microbiota dysbiosis and some bacteria are believed to induce the expression of genes related to lipid and carbohydrate metabolism [5]. For example, the relative ratio of Firmicutes/ Bacteroidetes in the host body is considered to be an effective indicator of whether the intestinal homeostasis is good [6]. It is reported that Firmicutes and Bacteroidetes contain nine genera of intestinal bacteria, such as Phascolarctobacterium, Faecalibacterium, Blautia, Bacteroides etc., produce Short-Chain Fatty Acids (SCFA). SCFA participate in part of the body’s total energy supply and disorder of its metabolic pathways can interfere with insulin signaling and thus preventing glucose metabolism [7-12]. Besides SCFA, Phascolarctobacterium can also be beneficial to human body thanks to its characteristics of succinic acid metabolism [7]. MiRNAs are a class of non-coding single-stranded molecules, their role in the regulation of T2DM at post-transcriptional gene level has been gradually demonstrated, which can be a therapeutic strategy to improve insulin resistance [4,13]. Interestingly, as early as 2016, Liu and his research team explicitly reported evidence that miRNAs synthesized by hosts can enter intestinal bacteria, complementary binding to 16S rRNA/23S rRNA sequences of bacteria and affect the growth of intestinal bacteria by regulating gene expression [14]. Perhaps there is a particular miRNA-mRNA tandem in the host-microbiome dysbiosis.
To deepen the understanding of the host-microbiome cross talk during the occurrence of T2DM and so as to obtain more potential intestinal microbial markers of disease, multi-omics analysis of human gut microbiota, which combined of microbiome, metabolomics, transcriptomics and so on is of considerable value in carrying out. Based on this, we designed a cohort study consisting of 60 urban adults in southern China, the volunteers were divided into a Normal Control (NC) (n=30, NC) group, a T2DM patient group (n=30, T2DM) and T2DM patients treated with metformin for 1 month (n=30) T2DM Metformin-therapy (TM), collected their feces and made 16S rRNA gene amplicon sequencing of gut microbiota. To explore different characters of intestinal bacteria and screen ideal microbial markers in T2DM, a multi-omics analysis of fecal microbiome, the results of metabolomics changes in host-microbiome dysbiosis and the candidate miRNAs genomics was conducted, combined with technologies such as bioinformatics prediction, Reverse Transcription Polymerase Chain Reaction (qRT-PCR), statistical methods of correlation and ROC curve analysis. Two early diagnostic models (ROC curves) for T2DM were constructed composed of candidate gut bacteria, fatty acids and blood miRNA from healthy people and T2DM patients.
Volunteers recruitment and samples collection
Volunteers of this study were recruited from the society and the Hospital of Chengdu University of Traditional Chinese Medicine from August 2020 to September 2020. All patients were diagnosed with early stage of T2DM, no complications associated with T2DM and no drug therapy before (the diagnostic criteria were based on WHO Diagnostic criteria for diabetes 1999). None of the subjects received any antibiotics or drugs designed to regulate gut flora. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Hospital of Chengdu University of TCM (2020KL-031). Patients who met the inclusion criteria were informed of the details of the research and signed an informed consent and we had access to information that could identify individual patients during data collection. All volunteers were divided into two groups; T2DM group consisting of 30 T2DM patients and normal control group consisting of 30 healthy individuals. T2DM patients had been told to receive metformin for one month and form TM group after treatment, their stool and blood samples were collected. In this study, stool samples were used for the 16S rRNA gene sequencing and blood samples were used for the detection of clinical diagnostic indicators of T2DM.
16S rRNA extraction and third-generation full-length sequencing
After pre-treatment, 500 mg of fecal sediment was added into a sterile Eppendorf tube. Total Deoxyribo-Nucleic Acid (DNA) of the sample was extracted using PowerSoil® kit (Qiagen, Hilden, Germany) as instructed. The corresponding full-length primer 27F-1492R was designed according to the conserved region for Polymerase Chain Reaction (PCR) amplification of the target region. PacBio Sequel for 16S rRNA sequencing was commissioned by Biotechnology company of BioMarker. The synthesized specific primers were designed as given below.
Forward primer 27F: 5′-AGRGTTTGATYNTGGCTCAG-3′
Reverse primer 1492R: 5′-TASGGHTACCTTGTTASGACTT-3′
The target region was then amplified with the KOD FX Neo amplification enzyme and the amplified products with sequence length of 1200-1650 base pairs were detected by gel electrophoresis. 1.8% agar gel was obtained by gluing, then the contents were poured into the gluing tank and the samples were prepared after cooling. 300 mg sample was added to each well in the Tanon HE- 120 electrophoresis tank and 3.5 μl marker was added to both sides of the sample. Ethidium Bromide (EB) staining was used for identification and Image J was used for quantification of final electrophoresis results. After terminal repair and the addition of a base to the obtained high-quality DNA fragments, the joint sequence was added to the end of fragments. Subsequently, fragment screening, sequencing primer hybridization and DNA polymerase binding were performed to construct the library. The Single Molecule Real Time (SMRT) chip was used as the sequencing carrier and PacBio Sequel for sequencing.
Total RNA extraction and qRT-PCR of miR-122-5p
The total RNA of plasma samples was first extracted using the QIAGEN miRNeasy Serum/Plasma (Qiagen, Hilden, Ger-many) extraction kit. The miRNAs were reverse-transcribed using Mir- X™miRNAs First-Strand Synthesis kit for total RNA, followed by real-time PCR using SYBR®qRT-PCR kit. In this study, U6 snRNA was used as reference gene and SYBR Green I chimeric fluorescence method was used to quantitatively analyze the synthesized complementary DNA, sequences of candidate miR- 122-5p and primer sequences (Table 1). By calculating the Cycle Threshold (Ct) value and fluorescence threshold, the relative quantitative determination of target gene sequences was realized.
| Gene | Sequence |
|---|---|
| miR-122-5p | 5′-UGGAGUGUGACAAUGGUGUUUG-3′ |
| miR-122-5p RT | 5′CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACACCA-3′ |
| miR-122-5p F | 5′-ACACTCCAGCTGGGTGGAGTGTGACA-3′ |
| miR-122-5p R | 5′-TGGTGTCGTGGAGTCGUGGUGUGUGUCUUUGG-3′ |
Table 1: Sequences of candidate miR-122-5p and primer sequences.
Statistical analysis
Gut microbiome diversity and difference significance analysis: The original data was filtered to obtain Circular Consensus Sequencing (CCS) sequence SMRT link (version8.0) after sequencing and UCHIME (version 8.1) was used for preliminary screening to remove chimeras and obtain high-quality optimized CCS sequence. USEARCH (version 10.0) was used to perform cluster analysis of the generated high-quality CCS at the similarity level of 97%. Corresponding species classification is obtained according to the Operational Taxonomic Unit (OTU) sequence composition. Species annotation and taxonomic analysis were carried out using 16S Silva database, Quantitative Insights Into Microbial Ecology (QIIME) software, R.Studio and other platforms and the diversity of intestinal flora was analyzed using Mothur version 1.30. To assess the difference of microbial community abundance between NC and T2DM samples, independent-sample t-test was performed on the species abundance of intestinal bacteria at the taxonomic level of genus and species using Metastats software and the species that caused the composition difference between the two groups were screened according to the p value (relabund P value<0.05) and then, the intestinal microbial biomarkers with statistical differences were screened by Linear Discriminant Analysis Effect Size (Lefse) analysis which performed in NC and T2DM groups (LDA score>4, P<0.05) and T2DM and TM groups (LDA score>3, P<0.05) respectively and the histogram of Linear Discriminant Analysis (LDA) value distribution was drawn. Random forest analysis was performed to construct prediction models at the species and genus levels (decision tree is 500) and the characteristic importance of the three groups of intestinal bacteria was evaluated. Other than that, all statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) 22.0 software (IBM Corporation, Armonk, New York, United States) or GraphPad Prism 7.0 (GraphPad Software, La Jolla, California, United States) with the adjust p value<0.05 was considered as statistically significant, notably, paired t test was used to analyze different intestinal bacteria between T2DM and TM groups.
Integrated analysis among host miRNAs gene and lipid metabolism dysregulations and changes in microbiome: For this analysis, Pearson analysis was used to analyze the correlation between Phascolarctobacterium and candidate miRNAs and Spearman analysis was used to analyze the correlation between Phascolarctobacterium/P. faecium and blood diagnostic indexes of T2DM. Subsequently, ROC analyze of bacteria with potential biomarker characteristics in combination with multiple parameters was performed, including the indicators of relative abundance of Phascolarctobacterium/P. faecium and α-linolenic acid. Semi-quantitative qRT-PCR data of miR-122-5p were analyzed to construct a joint prediction model for early diagnosis of T2DM.
Bioinformatics analysis
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software was used to predict the composition of functional genes in samples by comparing the species composition information obtained from 16S sequencing data. First, the generated OTU table was standardized and then, we got the corresponding Kyoto Encyclopedia of Genes and Genomes (KEGG) family information through the gene id corresponding to each OTU and the Pathway information was obtained from the KEGG database. In this study, the differences of KEGG metabolic pathway at level 3 were analyzed to observe the differences and changes of functional genes of microbial communities in metabolic pathway among samples of different groups, with an adjust P-value of 0.05.
The disorders of glucose and lipid metabolism in early stage of T2DM
There was no statistical difference in demographic information including gender and age distribution between two groups (P>0.05). T2DM patients showed abnormal glucose metabolism, lipid metabolism and insulin secretion (Table 2). Compared with healthy subjects, Body Mass Index (BMI), Fasting Plasma Glucose (FPG), Tri-Glyceride (TG), High-Density Lipoproteincholesterol (HDL), Low-Density Lipoproteincholesterol (LDL) and Fasting Serum Insulin (FINS) were significantly higher in T2DM patients (P<0.001, P<0.001, P=0.0125, P<0.001, P=0.022). Notably, TG is associated with insulin sensitivity, which is a risk indicator of T2DM, suggesting that lipid metabolism disorder represented by TG is correlated with the development of T2DM and HDL is significantly decreased in patients (P=0.0013) as a protective factor for cardiovascular disease, TG/HDL suggests that T2DM patients are at higher potential risk of cardiovascular diseases [15,16]. In addition, a metabolomics study in our research group recently also showed significant lipid metabolism abnormalities in patients with T2DM [17]. It can be seen that in the early stage of T2DM, metabolic disorders occur in the carbohydrate, lipid and insulin secretion in the host.
| Variable | NC (n=30) | T2DM (n=30) |
|---|---|---|
| Male (n/%) | n=13 | n=14(46.7) |
| Age (years) | 46.27 ± 8.44 | 48.13 ± 6.56 |
| BMI (kg/m²) | 22.37 ± 1.87 | 24.70 ± 3.00** |
| FPG (mmol/L) | 5.24 ± 0.69 | 7.52 ± 2.65** |
| HbAlc (%) | 5.45 ± 0.39 | 7.31 ± 2.06* |
| TC (mmol/L) | 4.63 ± 0.74 | 4.84 ± 0.66 |
| TG (mmol/L) | 1.28 ± 0.74 | 1.79 ± 1.02* |
| HDL (mmol/L) | 1.47 ± 0.39 | 1.18 ± 0.28* |
| LDL (mmol/L) | 2.68 ± 0.57 | 3.14 ± 0.66** |
| FINS (mmol/L) | 7.61 ± 2.76 | 10.41 ± 5.59* |
Note: All parameters are mean ± Standard Deviation (SD). Body Mass Index (BMI), Fasting Plasma Glucose (FPG), Hemoglobin A1c (HbA1c), Total Cholesterol (TC), Tri-Glyceride (TG), High-Density Lipoproteincholesterol (HDL), Low-Density Lipoproteincholesterol (LDL), Fasting Serum Insulin (FINS). ** adjusted P value<0.01; * adjusted P value<0.05.
Table 2: Demographic characteristics and baseline clinical features of NC and T2DM groups.
The disorders of glucose and lipid metabolism in early stage of T2DM
In this study, a total of 90 fecal specimens in three groups were sequenced by 16S rRNA full-length sequencing, which was different from second-generation sequencing, could identify the bacteria at species level. The original data showed that at least 428 CCS sequences were generated in each sample after the removal of chimeras and the effective rate was more than 77%. This indicated that the sequencing data was of good quality and can be further analyzed for species abundance and diversity. We found significant differences in intestinal microbial composition and abundance between healthy people and T2DM patients. The distribution of the top 10 intestinal flora and the bacteria among three groups were different at the phylum, genus and species levels (Figure 1A). Among them, Firmicutes, Bacteroidetes and Proteobacteria were the main dominant bacterial groups. Firmicutes and Bacteroidetes in the host have a symbiotic relationship and the proportion of them is considered to be an effective indicator to measure the stability of intestinal flora in body [6]. In this experiment, compared with NC group, the proportion of Firmicutes/Bacteroidetes in T2DM group increased from 0.93 to 1.73. This phenomenon may be related to the changes of metabolic pathways and phenotypes in multiple biological substrates, such as the decreased abundance of some butyrate producing probiotics and a variety of opportunistic bacteria may increase correspondingly [5,18]. At the genus and species level, we identified a total of 5 genera and 36 species of bacteria with significant differences by Metastats analysis (only the top 5 species in abundance were shown). To investigate the potential of the gut microbiota in distinguishing the T2DM patients from heathy people, Area Under the ROC Curve (AUC) values of the genera and species in LEfSe was further calculated. As the bar chat shows (Figure 1C), we identified a total of 7 microbial biomarkers between the two groups (biomarker screening criteria: LDA score>4, P<0.05). There are 4 types of intestinal bacteria significantly reduced in T2DM patients, including Phascolarctobacterium faecium, Phascolarctobacterium, Bacteriods stercoris, Bacteriods uniformis and while there are 3 types significantly increased, like Megamonas, Megamonas funiformis, Bacteriods coagulans. Among them, Phascolarctobacterium and P.faecium which belonging to the genus of Phascolarctobacterium had significant differences both in metastats and LEfSe analysis.
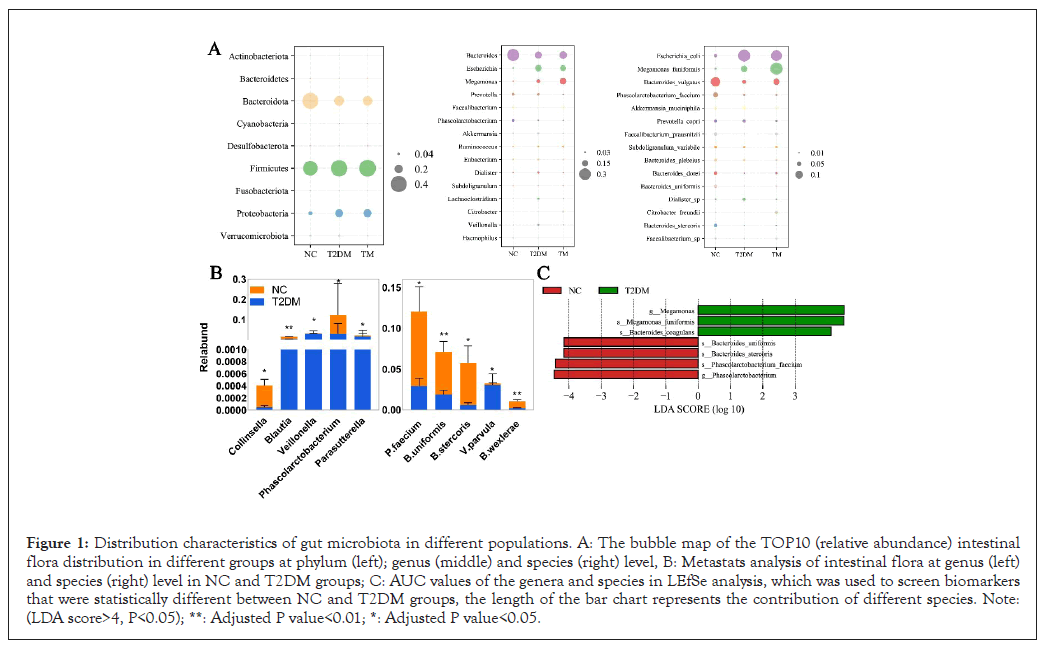
Figure 1: Distribution characteristics of gut microbiota in different populations. A: The bubble map of the TOP10 (relative abundance) intestinal flora distribution in different groups at phylum (left); genus (middle) and species (right) level, B: Metastats analysis of intestinal flora at genus (left) and species (right) level in NC and T2DM groups; C: AUC values of the genera and species in LEfSe analysis, which was used to screen biomarkers that were statistically different between NC and T2DM groups, the length of the bar chart represents the contribution of different species. Note: (LDA score>4, P<0.05); **: Adjusted P value<0.01; *: Adjusted P value<0.05.
Bioinformatics analysis and function prediction of intestinal microbial in T2DM
To observe the changes in metabolic pathways of functional genes in intestinal bacteria in patients with T2DM, we performed KEGG bioinformatics analysis for intestinal microorganisms in different groups. As shown, Adipocytokine signaling pathway (P=0.002), Peroxisome Proliferator-Activated Receptors (PPAR) signaling pathway (P=0.003), Taurine and hypotaurine metabolism (P=0.005) and other pathways were significantly reduced (Figure 2A). Adipocytokine is closely related to PPARs signaling pathway and thus involved in stable lipid metabolism in the body [19,20]. Many studies have confirmed the importance of taurine and hypotaurine in the treatment of diabetes [21,22]. And as our previous work showed that α-linolenic acid, which significantly decreased in T2DM had a positive correlation with Phascolarctobacterium [17]. Interestingly, α-linolenic acid can also regulate PPARs to improve metabolism and anti-inflammation [23,24]. Therefore, PPARs may be a crucial molecule in the regulation of host-microbiome metabolism at early T2DM, which deserves extensive discussion [25].
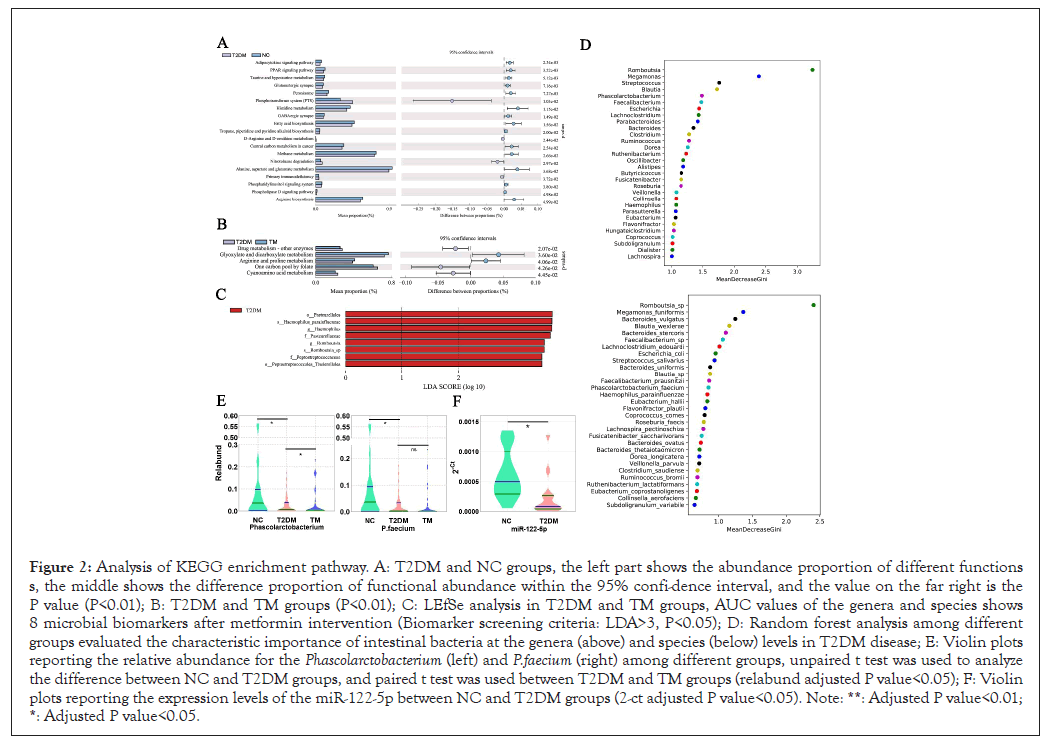
Figure 2: Analysis of KEGG enrichment pathway. A: T2DM and NC groups, the left part shows the abundance proportion of different functions s, the middle shows the difference proportion of functional abundance within the 95% confi-dence interval, and the value on the far right is the P value (P<0.01); B: T2DM and TM groups (P<0.01); C: LEfSe analysis in T2DM and TM groups, AUC values of the genera and species shows 8 microbial biomarkers after metformin intervention (Biomarker screening criteria: LDA>3, P<0.05); D: Random forest analysis among different groups evaluated the characteristic importance of intestinal bacteria at the genera (above) and species (below) levels in T2DM disease; E: Violin plots reporting the relative abundance for the Phascolarctobacterium (left) and P.faecium (right) among different groups, unpaired t test was used to analyze the difference between NC and T2DM groups, and paired t test was used between T2DM and TM groups (relabund adjusted P value<0.05); F: Violin plots reporting the expression levels of the miR-122-5p between NC and T2DM groups (2-ct adjusted P value<0.05). Note: **: Adjusted P value<0.01; *: Adjusted P value<0.05.
Besides maintaining stable glucose metabolism, metformin may have remodeling-implication of the gut microbiome
After 1 month of treatment with metformin, the clinical indexes of FPG and Hemoglobin A1c (HbAlc) in T2DM patients improved significantly (P<0.001, P=0.014, respectively). In terms of lipid, although there were differences in TC and TG, but with no statistical significance (Table 3). The possible reason is that the medication cycle is short and metformin needs a certain response period from simple control of blood sugar to effect on blood lipid and the same may be true for regulating insulin secretion [26].
| Variable | T2DM (n=30) | TM (n=30) |
|---|---|---|
| FPG (mmol/L) | 7.52 ± 2.65 | 6.21 ± 0.80** |
| HbAlc (%) | 7.31 ± 2.06 | 6.45 ± 0.9* |
| TC (mmol/L) | 4.84 ± 0.66 | 5.12 ± 3.34 |
| TG (mmol/L) | 1.79 ± 1.02 | 1.84 ± 1.37 |
| HDL (mmol/L) | 1.18 ± 0.28 | 1.11 ± 0.21 |
| LDL (mmol/L) | 3.14 ± 0.66 | 2.77 ± 0.86 |
| FINS (mmol/L) | 10.41 ± 5.59 | 9.80 ± 5.73 |
Note: All parameters are mean ± Standard Deviation (SD). Body Mass Index (BMI), Fasting Plasma Glucose (FPG), Hemoglobin A1c (HbA1c), Total Cholesterol (TC), Tri-Glyceride (TG), High-Density Lipoproteincholesterol (HDL), Low-Density Lipoproteincholesterol (LDL), Fasting Serum Insulin (FINS). ** adjusted P value<0.01; * adjusted P value<0.05.
Table 3: Baseline clinical features of T2DM and TM group.
Nevertheless, after metformin intervention, the composition of intestinal flora was partially changed. Compared with T2DM group, the proportion of Firmicutes/Bacteroidetes in TM group was increased (Figure 1A) and 8 microbial species were decreased after treatment (Figure 2C). The paired t test showed that Phascolarctobacterium increased in TM group (P=0.0456). Thus, Phascolarctobacterium were changed in both T2DM disease and after metformin treatment and the relative abundance of Phascolarctobacterium and P.faecium in different (Figure 2E). Subsequently, we predicted the metabolic pathway of intestinal bacteria in T2DM and TM group by KEGG analysis. As shown, there were statistically significant metabolic changes in drug metabolizing enzymes (P=0.021), glyoxylate and dicarboxylate acid (P=0.036) and the synthesis of arginine and proline metabolism (P=0.041) (Figure 2B). Glucose and amino acid metabolism are closely related through the tricarboxylic acid cycle, suggesting that one of the pathways of metformin in the treatment of T2DM may be related to the intervention of amino acid metabolism pathway in gut microbiome.
In addition, Phascolarctobacterium is a genus that grows with little use of carbohydrates but depends on succinic acid, which is a dicarboxylic acid [27]. Therefore, we have reason to conjecture that metformin is likely to ameliorate T2DM by affecting the Phascolarctobacterium and its metabolic pathway, while it is largely unknown and deserves extensive discussion. Hereafter, to assess the characteristic importance of intestinal bacteria at the genera and species levels between three groups, predictive models (decision tree is 500) were constructed by random forest analysis. MeanDecreaseGini is used to represent the importance of variables. Random forest analysis showed (Figure 2D) Phascolarctobacterium and P.faecium were both considerable for intestinal micro ecological imbalance in T2DM patients and whether they are ideal biomarkers for T2DM deserves further exploration.
Multi-omics analysis suggested that Phascolarctobacterium and P.faecium could be ideal markers of intestinal microorganisms of T2DM
As our previous association analysis between gut microbiome and miRNAs genomics, indicated that P.faecium was correlated with miR-122-5p [28]. In this study, correlation analysis also showed Phascolarctobacterium was correlated with miR-122-5p (Figure 3A). Given the previous high-throughput sequencing results of miRNAs, we then carried out qRT-PCR and found that the expression of miR-122-5p in T2DM patients was indeed increased by about 5.74 times (Figure 2F). MiR-122-5p is a valuable molecule in the occurrence and development of T2DM due to its regulatory effect on gluconeogenic genes like PPARs, PGC-1α and FOXO [29,30]. Thus, besides miR-122-5p itself as a good T2DM biomarker, association analysis with Phascolarctobacterium and P.faecium may provide the possibility for a better identification of potential molecular interactions in the host-microbiome during the disorder of glucose metabolism in the early stage of disease [31,32]. Furthermore, a total of 355 different small molecule metabolites were identified in T2DM by Liquid Chromatography Mass Spectrometry (LC-MS) in our previous study. Correlation analysis of fecal intestinal microbiome and metabolomics showed that Phascolarctobacterium was correlated with α-linolenic acid. In summary, the multi-omics joint of microbiome, metabolomics and miRNAs genomics indicated that Phascolarctobacterium and P.faecium could be considerable microbial markers for early diagnosis of T2DM in theory.
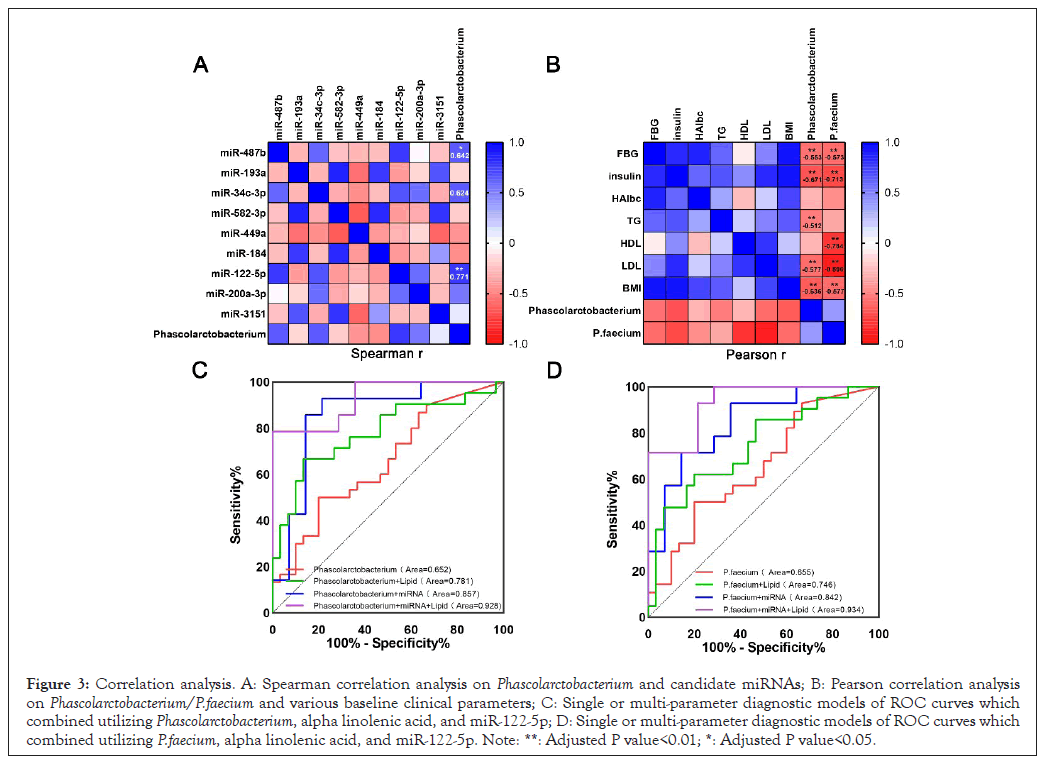
Figure 3: Correlation analysis. A: Spearman correlation analysis on Phascolarctobacterium and candidate miRNAs; B: Pearson correlation analysis on Phascolarctobacterium/P.faecium and various baseline clinical parameters; C: Single or multi-parameter diagnostic models of ROC curves which combined utilizing Phascolarctobacterium, alpha linolenic acid, and miR-122-5p; D: Single or multi-parameter diagnostic models of ROC curves which combined utilizing P.faecium, alpha linolenic acid, and miR-122-5p. Note: **: Adjusted P value<0.01; *: Adjusted P value<0.05.
ROC curves composed of Phascolarctobacterium/P. faecium, candidate miRNA and fatty acid are of great promising to improve the early diagnosis of T2DM
Spearman correlation analysis on Phascolarctobacterium/P.faecium and various baseline clinical parameters (Figure 3B) shows that P.faecium has a strong negative correlation with LDL (r=-0.89, P<0.001), HDL (r=-0.78, P<0.001) and FIN (r=-0.71, P<0.001) and also has a certain correlation with BMI (r=-0.57, P<0.001) and FBG (r=-0.58, P<0.001). Phascolarctobacterium has a certain correlation with FIN (r=-0.67, P<0.001) and BMI (r=-0.64, P<0.001).
Hereafter, we constructed two diagnostic models which combined utilizing data of Phascolarctobacterium/P.faecium, alpha linolenic acid and miR-122-5p (Figure 3C and 3D). ROC curves could classify T2DM patients from healthy people with the integrated analysis of the above three parameters accurately and the areas under the curve were 0.929 (95%Cl=0.84-1.00, p=0.0001) and 0.934 (95%Cl=0.85-1.00, p<0.0001) respectively. Notably, it is already comparatively accurate to combined Phascolarctobacterium/P. faecium and miR-122-5p for the diagnosis of T2DM, the areas under the curve were AUC=0.86 (95%Cl=0.70-1.00, p=0.001) and AUC=0.842 (95%Cl=0.70-0.99, p=0.002) respectively. However, only Phascolarctobacterium (AUC=0.652, 95%CI=0.51-0.79, p=0.042) or P.faecium (AUC=0.655, 95%CI=0.51-0.80, p=0.043) had low classification potential.
As a metabolic disease with long course and almost incurable, the diagnosis of T2DM is mainly based on blood glucose monitoring and observation of clinical signs. Thus, prevention and early diagnosis are currently the most effective health strategies for T2DM. In this population cohort-study, we characterized gut microbiota of stool samples by performing 16S rRNA gene amplicon sequencing. It is notable that we adopted the third-generation full-length sequencing method, which is different from the second-generation sequencing and can identify bacteria down to species-level. This is essential for further discussion of the causal relationship between gut flora and disease occurrence on the basis of their correlation and on the basis of previous studies, we conducted a multi-omics analysis of fecal gut microbiome, lipid metabolomics and blood miRNAs genomics between healthy people and T2DM patients. We observed metabolic disorders of lipid, carbohydrate and insulin secretion occurred in early T2DM hosts and related auxiliary diagnostic indicators were only partially improved after one month of metformin treatment. These results showed that one course of metformin treatment may not be enough to change the existing pathways of lipid metabolism and insulin secretion in the host and it is suggested that related studies should increase the trial period on this basis.
16S rRNA gene amplicon sequencing showed that T2DM patients had specific structural characteristics of intestinal flora. Similar to the discovery of previous studies, our work indicated the proportion of Firmicutes/Bacteroidetes changed after T2DM. SCFA and succinic acid metabolic disorders caused by the imbalance of the two ratios may be a possibility for the occurrence of metabolic diseases [5,7]. Analysis of significant difference between the groups showed that the relative abundance of Phascolarctobacterium and P.faecium decreased in T2DM patients, both of which are negatively correlated with insulin secretion and obesity and P.faecium has a certain negative correlation with glycolipid metabolism. Metformin treatment had a remodeling-implication of the intestinal micro ecology and the relative abundance of Phascolarctobacterium increased. Bioinformatics predictions suggested that metformin may ameliorate T2DM by affecting the relative abundance of Phascolarctobacterium and its metabolic pathway, which is worth extensive discussion.
Phascolarctobacterium and P.faecium have colonized in the gastrointestinal tract of human beings and belong to Firmicutes with the largest number in gut microbiota, their abundance is relatively higher in Asian people [33]. A population study in southern China, which collected fresh feces from 150 subjects, found that the colonization rate of P.faecium in different populations ranged from 43.33 to 93.33% and the abundance was about 3.22-5.76 log cells/g (<1 year old) and 3.06-9.33 log cells/g (>1 year old). With the increase of age (1 to 60 years old), the number of bacteria gradually increased, but there was a decrease in older people (>60 years old) [27].
As one of the main strains of Phascolarctobacterium, P.faecium was first isolated from koala feces [34]. Recently, scientists successfully extracted P.faecium JCM 30894 from feces of healthy Japanese men and completed the genome sequencing [35]. The successful extraction and culture of P.faecium from feces provided a favorable way for further research. P.faecium can participate in host metabolism through the production of SCFA, it hardly use carbohydrates to grow, but succinic acid is essential for its reproduction. Since succinic acid is a key metabolite for the growth of Clostridium difficile, the enrichment of P.faecium can prevent the growth of Clostridium difficile [33]. In addition, the excessive accumulation of succinic acid in the host will lead to diarrhea, thus P.faecium may be beneficial to human body since its properties of utilizing succinic acid and producing SCFA. Phascolarctobacterium is negatively correlated with the stability of lipid and mass metabolism in human obesity [36-39]. A Mayo Clinic center of obesity treatment shows some significant differences in gut bacteria between people who failed to lose weight and those who succeed. People who lose weight easily have higher levels of Phascolarctobacterium in their gut, while the level of Dialister is lower [40]. It is also reported that leptin and insulin sensitivity were associated with higher abundance of Phascolarctobacterium and lower abundance of Dialister [37,39]. A population study on urban adults in China and rat experiment both suggested that Phascolarctobacterium may be related to the prevention of nonalcoholic fatty liver disease [38,41]. Furthermore, a study conducted 16S rRNA sequencing of gut microbiota on T2DM macaque feces showed that the abundance of Phascolarctobacterium producing short-chain fatty acids significantly decreased in disease [42].
Therefore, accumulating evidence from reports in multiple experimental disease, models indicates that Phascolarctobacterium is closely related to lipid metabolism and insulin sensitivity, it is of great importance for glycolipid metabolism in early stage of T2DM diabetes, but the target molecules of Phascolarctobacterium with functional activities in the host-microbiome have not been widely discussed. Moreover, similar to the discovery of previous studies, our work showed that metformin, as an effective drug for T2DM, has recovery effect on intestinal microbiome disorder and can significantly increase the abundance of Phascolarctobacterium [43,44]. That also means the existence of Phascolarctobacterium may contribute to the beneficial regulation of metformin on host sugar metabolism disorder in some degree. To assess the ability of Phascolarctobacterium and P.faecium as biomarkers for clinical diagnosis, we identified two diagnostic models of ROC curves for T2DM. ROC curves composed of Phascolarctobacterium/P.faecium, α-linolenic acid, miR-122-5p could classify T2DM patients from healthy people accurately.
In view of the characteristics and importance assessment of gut microbiota and the multi-omics analysis between different groups, our study proposed firstly that Phascolarctobacterium and P.faecium are promising biomarkers of T2DM. They are core intestinal bacteria in host-microbiome metabolism and their beneficial effects may be closely related to the characteristics of SCFA and succinic acid metabolism, which can be the basis for further mechanistic studies and clinical tests. Due to considerable progress has been made in the application of single-cell sequencing, Virus transfection and LC-MS technologies, the identification and validation of miRNAs and their target genes, as well as small molecule metabolites have shown promising aspects in the predicting of T2DM occurrence and development. However, there is still insufficient on the research methods and technologies to uncover the underlying mechanism of target molecules in the occurrence of diseases caused by gut microbiota. It is hoped that more multi-omics joint studies will be carried out to screen for more microbial metabolites and determine how they interact with the host targets in improving host metabolism, which is also considerable to improve early clinical diagnosis of T2DM.
Lisha Li, methodology, software, visualization, validation, formal analysis, investigation and writing—original draft preparation; Qiongying Hu, resources, data curation, writing—review and editing, supervision; Daqian Xiong, writing review and editing, supervision, project administration and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Daqian Xiong: Key Research and Development Program of Sichuan Province, China. (2020YFS0375)
I would like to thank to the Department of Endocrinology and Physical Examination of the Hospital of Chengdu University of Traditional Chinese Medicine for their support of this volunteer recruitment.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Li L, Hu Q, Xiong D (2023) Associations between Phascolarctobacterium/Phascolarctobacterium faecium and Disorders of Glucose and Lipid Metabolism in Type 2 Diabetes Mellitus-A Case Control Study. J Proteomics Bioinform. 16:655
Received: 01-Nov-2023, Manuscript No. JPB-23-27817; Editor assigned: 03-Nov-2023, Pre QC No. JPB-23-27817 (PQ); Reviewed: 20-Nov-2023, QC No. JPB-23-27817; Revised: 27-Nov-2023, Manuscript No. JPB-23-27817 (R); Published: 04-Dec-2023 , DOI: 10.35248/0974-276X.23.16.655
Copyright: © 2023 Li L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.