Journal of Glycobiology
Open Access
ISSN: 2168-958X
ISSN: 2168-958X
Research Article - (2018) Volume 7, Issue 2
The glycosylation state of therapeutic antibodies is a critical quality attribute due to its effect on product efficacy and safety. With the advent of the biosimilar development and the need to match the glycan profile of the originator product, better understanding of how the variables of the bioprocessing procedure control glycosylation is critical. To this effect, we used automated microbioreactors with our in-house model CHO DG 44 cell line and different media types to study differences in antibody outcomes, specifically focusing on N-glycosylation profiles and aglycosylation rates. We observed that different media types resulted in vastly different amounts of high mannose and terminal galactosylation of N-glycans. By measuring the percentage of antibodies that was not N-glycosylated we observed that high mannose outcomes were not correlated to aglycosylation rates. For further analysis, we utilized multivariate data analysis to determine the process variables that best explained our glycan profile findings. Factors linked to glutamine consumption were determined to be the most important in predicting high mannose outcomes, while factors related to the temporal aspects of cell growth rate were linked to terminal galactosylation. Our work discovered in-process parameters in the cell culture process that have significant effects on the glycan profile of an antibody product, further elucidating the link between the biomanufacturing process and product quality outcomes.
Keywords: Bioprocessing; Glycosylation; MVDA; PCA; Microbioreactors; Media; Glutamine; CHO
Chinese hamster ovary (CHO), Monoclonal antibody (mAb), Critical Quality Attribute (CQA), Multivariate data analysis (MVDA), Complement-dependent cytotoxicity (CDC), endoplasmic reticulum (ER), N-acetylglucosamine (GlcNAc), crystallizable fragment (Fc), Antibody-dependent cellular cytotoxicity (ADCC), Principal Component Analysis (PCA), Partial Least Squares (PLS), Harvest cell culture fluid (HCCF), Dimethylformamide (DMF), acetonitrile (ACN), Similar Integrated Viable Cell Density (IVCD), High Mannose (HM), Mannose 4 (M4), Mannose 5 (M5), Mannose 9 (M9).
Nutrient-rich bioreactor media is essential for optimal production from cultured Chinese hamster ovary (CHO) cells that have become the preferred platform for the manufacturing of therapeutic proteins such as monoclonal antibodies (mAb) [1,2]. The starting concentrations of these nutrients are critical, especially under culturing conditions such as in batch mode where they are not replenished over the course of the production run. Depletion of nutrients has serious impacts on the product produced, including the amounts and Critical Quality Attributes (CQA) such as product glycosylation in mAbs [3]. We used the ambr®15 microbioreactor system and multivariate data analysis (MVDA) to determine cell culture parameters that have significant impacts on product quality, focusing on N-glycosylation of a model mAb. This includes differences in bioreactor media which altered product terminal galactosylation and the abundance of high-mannose N-glycans, which affect complement-dependent cytotoxicity (CDC) and product half-life, respectively [4]. N-glycans on IgG mAbs are assembled on the protein in the same fashion as other glycoproteins: briefly, a 14-sugar precursor Glc3Man9GlcNAc2- is attached to Asn297 of the IgG heavy chain in the endoplasmic reticulum (ER) and is further processed as it passes through the cis, mid- and trans-Golgi apparatus [5]. As the mAb traverses through the Golgi, most mannose residues are typically trimmed off and replaced with N-acetylglucosamine (GlcNAc) which is further modified by galactose residues. As these processes require enzymatic activity, cellular stresses and nutrient shortages can affect modification rates.
Bioreactor parameters have significant effects on the quantity and quality of the produced protein product [6,7]. In the case of mAb products, one particularly important CQA is the distribution of product glycoforms [8,9]. N-linked glycosylation affects the therapeutic properties of the antibody, from effector function, immunogenicity, stability, to clearance rate [10]. IgG antibodies feature conserved N-glycosylation sites at Asn297 in the crystallizable fragment (Fc) of heavy chains, which influence effector binding to downstream molecules such as Fcγ receptors that mediate antibody-dependent cellular cytotoxicity (ADCC), a critical mechanism for the efficacy of many biotherapeutics [11]. A comprehensive understanding of the glycan profile of an antibody product and the nutrient parameters that generate the desired profile could be of great value for the development of biosimilars, where matching the proportions of glycoforms in the originator product is of critical importance [5].
Replicating the glycan profile of the originator product is important for biosimilars due to its effects on efficacy and product half-life. The mechanisms by which different glycoforms affect the performance of the antibody product has been well characterized, such as core fucose and how its presence sterically inhibits interaction with the Fcγ receptor and reduces ADCC-mediated activity [12]. Conversely, the factors in the growth and culture conditions that have substantial effects on the glycan profile are not well understood. Furthermore, the role of the nutrients in the media such as sugars, amino acids, and metals have been only minimally characterized with respect to how they can affect protein production and efficacy [13].
In addition to aberrant glycan profiles, complete lack of N-glycosylation (aglycosylation) is a possible outcome as well and can have dramatic effects on product efficacy depending on the mechanism of action of the therapeutic [14]. Notably, these aglycosylated antibodies cannot recruit and activate immune effector cells critical for ADCC. The lack of glycosylation also impacts the overall stability [4,15], propensity for aggregation at low pH [16], pharmacokinetic properties [17], and susceptibility to protease activity [15]. Complementary to this, IgG1 antibodies lacking an N-linked glycan have been investigated for targeting strategies that do not require Fcγ binding, and in these cases it can be preferable to mutate Asn297 to block glycosylation altogether [14]. Clearly, understanding the cell culture parameters to control aglycosylation can have beneficial effects such as increasing the half-life of the therapeutic antibody and preventing aggregation.
To promptly test a wide array of media conditions with sufficient simultaneous replicates we used automated microbioreactors, namely the ambr®15 (Sartorius, Hartfordshire, UK) system. In this study an inhouse model IgG1 producing CHO DG44 cell line was used. The culture conditions included four commercial media: ProCHO5, PowerCHO2, Ex-Cell Advanced and OptiCHO that had various culture supplements: glutamine, vitamins and essential amino acids. A range of working cell culture conditions were evaluated with glutamine, vitamins and essential amino acids as the variables. The media were selected based on prior studies [18]. After obtaining the purified antibody product, this material was characterized for glycan profile and size variants using mass spectrometry and capillary electrophoresis, respectively. Through MVDA, we linked bioreactor process parameters with increased efficiency in minimizing high mannose glycoform formation and altered proportions of terminal galactosylation. Our results demonstrate the wide range of product quality outcomes possible and the strong influence that media components have on the final glycosylation state of the produced mAbs, potentially allowing for real-time corrections to manage the glycan profile.
Cell culture reagents
Cells were originally cultured in CD OptiCHO media (Thermo Fisher Scientific, Waltham, MA). The cells were then additionally adapted into three other commercially available media: ProCHO5 (Lonza, Walkersville, MD), PowerCHO2 (Lonza, Walkersville, MD), CD Ex-Cell Advanced (SAFC, Lenexa, KS). Standard seed train protocol was used to inoculate the microbioreactors [19]. No supplements were added to the media during the seed train phase except for 8mM glutamine (Corning, Manassas, VA) and 1X Penicillin/Streptomycin (Corning, Oneonta, NY). The supplements were added to the media when inoculating the microbioreactors. The culture supplements used were Glutamine (200 mM), Essential Amino Acids (50X) (EAA) (Thermo Fisher Scientific, Grand Island, NY) and Vitamins (100X) (Thermo Fisher Scientific, Grand Island, NY).
AMBR cell culturing
All cultures were run in batch mode in an ambr®15 system (Sartorius, Hertfordshire, UK) containing four culture stations consisting of 48 disposable bioreactors (only sparged). Set points for process parameters for all reactor vessels were the same with dissolved oxygen (DO) at 50%, pH at 7.1 ± 0.05, temperature at 37°C and agitation rate at 1000 rpm. The reactor vessels were charged with media, inoculated, sampled and had additions with the use of the liquid handler. The vessels were inoculated with CHO DG44, an in-house cell line, at a density of 1 × 106 cells/mL and the culture volume was always maintained above 10mL and below 16 mL. Total cell density and viable cell density was measured using a Vi-Cell XR cell viability analyzer (Beckman Coulter, Brea, CA). Daily nutrient values were measured using a BioProfile Flex Analyzer (Nova Biomedical, Waltham, MA). The pH was maintained using in-house CO2 and 1 M NaOH (Thermo Fisher Scientific, Fair Lawn, NJ) [19]. The amount of CO2 input is managed by the system based on the pH.
Cell culture data preprocessing
All data preprocessing and parameter calculations were performed in MATLAB R2017 (Mathworks, Natick, MA).
Due to the potential for measurement error as high as 10% for the Vi-Cell XR or as high as 30% for the Bioprofile Flex Analyzer, it was important to mitigate its impact on subsequent calculations and analyses. This was accomplished using the Shape Language Modeling toolbox to fit a smooth profile to each of the measured time-series using a cubic interpolating spline with six knots [20].
The smoothed profiles were used to derive auxiliary parameters that could be correlated with product quality. Furthermore, the splines were used to estimate parameter values at 0.25 day intervals so that accurate numerical derivative and integration techniques could be employed for their calculation. Integrals were calculated using a Newton-Cotes integration formula for 5 points, as shown in equation θ1 where a and b are the integration boundaries, h is the step size defined by 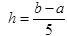 and
and 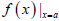 is the parameter estimate when the spline is evaluated at x = a [21].
is the parameter estimate when the spline is evaluated at x = a [21].
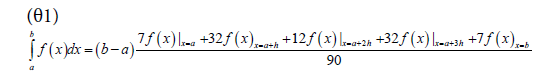
Due to the uneven spacing of some independent variables, the derivatives were evaluated from a 2nd order Lagrange polynomial fit to the data, as shown in equation θ2. It uses three consecutive measurement pairs (x0, f(x0)), (x1, f(x1)), (x2, f(x2)) to evaluate the derivative at some point x ϵ (x0, x2) [21]. For the first time point, x = x0. For all internal data points x = x1 and for the final time point x = x2.
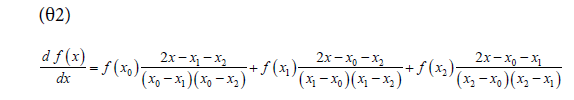
Using the above formulae, the growth rate and integrated viable cell density profiles are both calculated from the viable cell density profiles and their averages for each media type are shown in Figure 1A. In addition, nutrient consumption and metabolic byproduct formation rates, with respect to time and cell density, were calculated and are shown in Figure 1B. Furthermore, the amount of each nutrient consumed and byproduct formed were calculated for each 24-h interval and the cumulative total amounts were calculated as well, the latter being shown in Figure 1B. The end result is 5 measurements and 15 calculated parameters that can be correlated with media type and glycan profile.
Figure 1: Bioreactor growth profiles and nutrient profiles averaged across media type (A) Average Viability, Viable Cell Density, Growth Rate and Integrated Viable Cell Density across all culture conditions for ProCHO5, PowerCHO2, Ex-Cell Advanced and OptiCHO media. (B) Average glucose, glutamine consumption and lactate production across all culture conditions for ProCHO5, PowerCHO2, Ex-Cell Advanced and OptiCHO media in terms of nutrient and metabolite concentration, time derivative, cell derivative and total change per cell.
Multivariate data analysis
In exploratory data analysis, Principal Component Analysis (PCA) is used to make the data easier to visually interpret. This is possible because the measured variables, collectively referred to as the feature space, vary collinearly with one another; for example, an increase in the percent abundance of one glycoform must result in an equal cumulative decrease in the percent abundance of the other glycoforms. PCA exploits this collinearity by projecting the feature space onto a small number of latent variables, called principal components, that describe the variability in the original feature space [22]. Observations that are similar across many of the original variables will appear clustered together in the projection, referred to as the score space. Therefore, any observations, or groups of observations, that deviate from the others can be seen. PCA is used to establish the difference in glycan profiles that exists in the protein produced by cell cultures that use different media. The relationship between the feature space, X, and the score space, t, is described by equation θ3. The loadings, p, describe the relative orientation of the score space to the feature space. The residual matrix E contains the feature space’s variability that isn’t characterized by the A extracted principal components, where A is determined by 7-fold cross-validation to prevent model over-fitting [23].
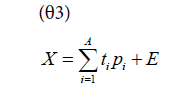
Partial Least Squares regressions (PLS) are used to create a regression model between two blocks of data that have been projected into the score space [24]. This allows for more robust regression models to be built due to the uncorrelated noise in the raw data being left out of the projection to the score space. PLS is used to establish the dependence of various glycoforms on in-process measurements obtained during cell culture and to establish which parameters are correlated with which glycoforms and for determining how strong that correlation is.
Monoclonal antibody purification
The harvest cell culture fluid (HCCF) was sterile filtered using a 0.22 μm PVDF membrane. The sterile HCCF was then purified using an AKTA Avant Protein Purification System (GE Healthcare Life Sciences, Pittsburg, PA). The stationary phase used was ProSEP® vA Ultra resin packed to 10 cm into an Omnifit (Diba Fluid Intelligence, Danbury, CT) glass housing with 0.66 m diameter. The equilibration buffer used for the in-house antibody was 25 mM Tris in 100 mM NaCl at pH 7.5 and 0.1 M Acetic Acid was used as the elution buffer. The high salt wash buffer used was 25 mM Tris with 1M salt at pH 7.5. All Tris buffers were prepared from 1M Tris-Base stock solution (Alfa Aesar, Ward Hill, MA) and NaCl (Thermo Fisher Scientific, Fair Lawn, NJ). The sample was injected manually using a 10 mL superloop system (GE Healthcare Life Sciences, Pittsburg, PA). The purified protein sample concentration was measured using the NanoDrop One microvolume UV-Vis spectrophotometer (Thermo Fisher Scientific, Fair Lawn, NJ) [19].
Concentration of purified antibody
The purified antibody was concentrated using Amicon 100 kDa filters. To remove glycerin, the filters were rinsed with 500 μL of 0.1 M NaOH and centrifuged for 10 min at 14,000 xg at room temperature. The filters were washed with 500 μL of 18 MΩ water and centrifuged for 10 min at 14,000 xg. This wash step was repeated a total of three times. The filters were transferred to fresh collection centrifuge tubes, and 500 μL of sample was centrifuged for 10 min at 14,000 xg. The filters were inverted into fresh collection tubes and centrifuged for 2 min at 1,000 xg to collect the concentrated sample. The sample concentration was determined using a Thermo Scientific NanoDrop One microvolume UV-Vis spectrophotometer. The spectrophotometer was blanked using the solution of 0.1 M Acetic Acid neutralized to pH 5.5 with Tris Base (Sample Buffer). A protein extinction coefficient of 13.7 at 280 nm for a 1% IgG solution was used in the calculation of the antibody concentration.
Glycan characterization
For glycan identification by LC-MS, the purified antibodies were processed by the Waters GlycoWorks platform for the deglycosylation, labeling and isolation of the glycans. Starting concentrations of 2 mg/ mL of antibody were used, diluted in equilibration buffer from the purification step. We followed the protocol provided by Waters without any deviations: Briefly, 7.5 μL of the antibodies were diluted with LCMS- grade water and then denatured using 5% RapiGest SF at 90°C for 3 minutes. Intact mAb Mass Check Standard was processed alongside experimental samples as a positive control. After cooling to room temperature, Rapid PNGase F was added to the denatured antibodies to cleave the glycans from the antibodies for 5 min at 50°C.
After cooling to room temperature, the cleaved glycans were labeled with RapiFlour-MS reagent dissolved in anhydrous dimethylformamide (DMF). The labeling reaction occurred at room temperature for 5 min. The labeled glycan mixture was diluted with acetonitrile (ACN) prior to clean-up.
The GlycoWorks HILIC μElution Plate was used with the vacuum manifold. The plate was conditioned with water, equilibrated with 85% ACN prior to loading the ACN-diluted labeled glycan mixture.
The vacuum was adjusted so the liquid would take from 15-30 sec to pass through the HILIC resin. The resin was washed with 1% formic acid/90% ACN twice before eluting with SPE elution buffer (3 elutions of 30 μL each). The elutions were pooled before diluting with DMF/ ACN to be ready for FLR-MS analysis. These samples are stable when stored at -80°C for at least 1 month.
The labeled glycan elution samples were run on a Waters ACQUITY I-Class UPLC coupled to a ACQ FLR fluorescence detector and Xevo G2 Q-ToF. The ACQUITY UPLC Glycan BEH Amide (130 Å, 1.7 μm, 2.1 × 150 mm) column was used for chromatographic separation of the labeled glycans, which was heated to 60°C during operation. The mobile phases were 50 mM ammonium formate (used of Waters mobile phase concentrate) and 100% LC-MS-grade ACN. The flow rate is 0.4 mL/min, with the LC gradient as provided in the GlycoWorks Care and Use Manual. The FLR measured at EX 265/EM 425 nm with a sampling rate of 2 Hz. The Q-ToF was operated in MS1 positive ion sensitivity mode, with a mass range of 100-2000 Da, a scan time of 0.25 sec and continuum data acquisition. Sodium iodide was used for the LockSpray, in the “Do NOT apply correction” mode.
Labeled glycan samples were run in triplicate, with error bars indicating standard deviations. RapiFlour-MS Dextran Calibration Ladder was used to determine identities of the glycans, with the mass information used to validate identifications. Data processing was performed using Waters UNIFI Scientific Information System optimized to run HILIC-FLR-MS data. LockSpray corrections were applied in UNIFI.
Non-glycosylated heavy chain
The percentage of non-glycosylated heavy chain of purified antibody was determined by rCE-SDS using the GXII HT Touch micro capillary electrophoresis system (Perkin Elmer) with a Protein Express Assay LabChip (PerkinElmer, cat#760499) and complementary Protein Clear HR Reagent Kit. LabChip and sample preparation were performed per the manufactures’ protocol. Briefly, Protein A purified samples were prepared by diluting each sample to 1 mg/mL in storage buffer, mixing 2.5 μL of sample with sample buffer supplemented with 8.75 mM DTT, heat denaturing the samples at 70°C for 10 min, and then adding 35 μL of water. The system was calibrated using the VeriMAb Standard and samples were then analyzed in triplicate. Electropherograms were exported to Empower 3 FR2 for peak integration, using the Gaussian skim and shoulder detect features. The size of the non-glycosylated heavy chain was verified by treating the antibody with PNGase F (Promega, cat#V4483A) and monitoring the shift in peak location from glycosylated to non-glycosylated heavy chain with and without PNGase F treatment, respectively (data not shown).
Four commercially available media (ProCHO5, PowerCHO2, Ex-Cell Advanced and OptiCHO) were used to examine the effects of media on glycosylation patterns of our model IgG1 antibody. All four media were inoculated using our in-house CHO DG44 cell line after adaptation to the different media. The cells were inoculated at a density ~1 x 106 cells/mL. The average growth and viability profiles of the cells cultured in these media are shown in Figure 1A. It can be seen from Figure 1A that while across all media types the viability remained constant, the growth rate and peak viable cell density differed with OptiCHO and Ex-Cell Advanced having the highest peak viable cell density and growth rate followed by ProCHO5 and PowerCHO2. (The average growth rate and viable cell density profiles were similar across different culture conditions in a media and were not significantly different). Similar Integrated Viable Cell Density (IVCD) was seen across all media except for OptiCHO, which had the highest IVCD. The specific productivity of the cultures ranged between 1.2-2.5 pg/cell-d and no differences were observed across different media groups.
A Bioprofile Flex Analyzer was used to measure the consumption of glucose and glutamine and the production of lactate as seen in Figure 1B. Glucose levels varied with media with ProCHO5 having the highest starting glucose level at ~8g/L and OptiCHO having the lowest at ~5g/L, while initial glutamine concentration for all media was at 8mM. Rate of glucose and glutamine consumption were similar across all media. Rate of lactate production was similar across all media with the exception of OptiCHO which had a lower rate of lactate production compared to other media. Though there were no apparent differences in growth, nutrient and metabolite profiles across media we reasoned that they could have an effect on the glycan profiles of the antibody therefore we performed Multi-Variate Data Modeling (MVDA) to analyze effects not observed with univariate analysis.
A significant factor affecting the product glycan profile was the type of media used in the microbioreactor vessels, as illustrated in Figure 2. Figure 2 depicts the average glycan percentages that were obtained with the all the nutrient conditions for each media tested, three replicates each. The glycan categories that we used for this study are as follows: G0F, G1F, G2F, High Mannose (HM) (which includes all glycoforms that primarily consist of mannose oligosaccharides bound to the 2 GlcNAc core from Mannose 4 (M4) through Mannose 9 (M9), and Other (which contains all the remaining glycoforms such as G0, G0F-N, G1 and G2). The use of these categories allowed us to more easily visualize and analyze the data to determine significant trends, as the “Other” grouping contained less than 5% of the total overall glycan profile. The ratio of terminal galactosylated species (G1F and G2F versus G0F) varied greatly based on media type: in ProCHO5 there is more G1F produced than G0F, while in OptiCHO there was more G0F produced than the sum of G1F and G2F combined. The ratios of terminal galactosylation, (G1F+G2F)/G0F, for each media type were as follows: Ex-Cell Advanced=1.00, OptiCHO=0.61, PowerCHO2=0.83, ProCHO5=1.50. The terminal galactosylation across the different media types vary significantly, as OptiCHO has low terminal galactosylation (0.61) while ProCHO5 has high terminal galactosylation (1.50) signifying that OptiCHO results in approximately 2.5 times more terminal galactosylation than ProCHO5. As glycan processing involving galactosylation requires more reactions to produce G1F and G2F when compared to G0F [25], this indicates that trafficking through the Golgi apparatus occurred optimally in ProCHO5 media.
Figure 2: Media dependent changes in glycan profiles Glycan profiles categorized by media type: all samples for each media type were combined for the data displayed. Glycans were divided into the following categories: G0F, G1F, G2F, High Mannose, and Other. Error bars represent the standard deviation of all the data points within each media type. The (G1F+G2F)/G0F numerical values underneath the graph represent the terminal galactosylation ratio: the sum of the percentages of the G1F and G2F values divided by the G0F value. Higher values indicate more terminal galactosylation. The (G0F+G1F+G2F)/HM values reflect the amount of high mannose species relative to the total distribution, with higher values indicating less of the high mannose forms.
The abundance of high mannose glycoforms was also linked to media type. Mannose 5 (M5) is a common example of a high mannose glycoform [26]. These high mannose glycans typically result from incomplete processing during N-glycan biosynthesis, and are indicative of cellular stress that can occur due to nutrient depletion or culture age [4,27]. As a score for high mannose glycoforms relative to the other forms in each media type, we used the sum of the values for the fucosylated species divided by the abundance of high mannose forms: (G0F+G1F+G2F)/HM (Figure 2). The bioreactors containing ProCHO5 contained significantly less high mannose forms than any of the other reaction vessels, as reflected by the high value of (G0F+G1F+G2F)/ HM equal to 16.35. Together with the terminal galactosylation results, this would indicate that our cell line responds with increased glycan trafficking when in ProCHO5 media than in any other type of media tested. This is significant as it indicates that significantly more terminal galactosylation can be achieved by choosing the correct media.
The results of the media-dependent glycan profiles can be combined with aglycosylation percentages for a more comprehensive understanding of how the mAbs are affected by media conditions (Figure 3). We observed that three of the four media types resulted in around 4.5% aglycosylation, but one media type, PowerCHO2, had almost double the amount of mAb aglycosylation at 8.32%. Interestingly, while PowerCHO2 raised mAbs featured the highest amount of aglycosylated mAbs, this media type was not the one that resulted in the largest percentage amount of high mannose forms or the lowest amount of terminal galactosylation. As determined earlier, the terminal galactosylation ratio for PowerCHO2 was 0.83, which was close to the average ratio for all media types (0.985), and the percentage of high mannose forms was second highest at 13.73%.
Figure 3: Media dependent changes in non-glycosylated heavy chain The percentage of non-glycosylated heavy chain categorized by media type: nutrient mid-points for each media type are displayed. Values are the average of three experimental replicates of each nutrient mid-point condition, with the error bars representing the standard deviation of the three replicates (except Ex-Cell Advanced, N=2).
Figures 2 and 3 together show that the glycan outcomes of the produced mAbs vary significantly with media type. While this is reflected in the glycan profile in terms of terminal galactosylation ratio and amount of high mannose forms, we discovered that aglycosylation was not directly linked to either poor terminal galactosylation or large values of high mannose form glycans. This is significant because one might assume that nutrient starvation conditions that result in insufficient processing in the Golgi and increased amounts of high mannose N-glycans could also disrupt the enzymatic pathways that are responsible for initially attaching the N-glycans to the mAbs, but our data supports that this is not the case. PowerCHO2 was determined to cause significantly higher amounts of aglycosylation, but PowerCHO2 is not the media where the largest amounts of high mannose forms or the lowest ratio of product terminal galactosylation were observed. These results would indicate that these outcomes are not necessarily linked directly, and therefore careful control of the glycan profile outcome might not minimize aglycosylation in the final product concurrently. Our work demonstrates the importance of fully characterizing the mAb product to maximize process-dependent product quality outcomes.
Principal Component Analysis (PCA) was performed to assess the suitability of using multivariate data analysis (MVDA) when characterizing the impact of media selection on antibody glycosylation. The glycosylation profiles alone were used as the model’s features; media type itself was not included in the model. The resulting model is summarized in Table 1 and the score space and loadings plot are shown in Figure 4. The two extracted principal components characterize approximately 95% of the glycosylation profiles’ variability. Furthermore, the high Q2 value indicates that the results are not based on spurious correlations and should be reproducible.
Figure 4: Principal Component Analysis model for the glycosylation profiles The score space shows that when the glycosylation profiles are projected onto principal components one and two, the observations are clustered according to media type. The loadings plot indicates that cells grown in EX-Cell Advanced produce a large proportion of high mannose glycoforms and that those grown in ProCHO5 feature a large degree of terminal galactosylation. Cells grown in OptiCHO and PowerCHO2 are the intermediates, as they are able produce protein with a large degree of the G0F glycoform, but less of the G1F and G2F glycoforms.
| A | R2 | Q2 |
|---|---|---|
| 2 | 0.943 | 0.734 |
Table 1: Glycosylation Profiles’ Model’s Summary Statistics
The score space in Figure 4 shows four distinct clusters in the score space, each of which corresponds to one media type. Furthermore, it can be seen from the loadings plot in Figure 4 that the first principal component primarily characterizes the exchange of high mannose glycoforms for G1F and G2F glycoforms and the second principal component characterizes the exchange of the G0F glycoform for all the others. The “Other” glycoform category was not used for PCA and MVDA analysis.
In considering the score space and loadings plot simultaneously, it is possible to assess the impact of media selection on glycosylation directly; proceeding in a counter-clockwise direction from the bottom right quadrant one can see that the principal components directly characterize the metabolic efficiency of the cell cultures grown in the four different media types. In the bottom-right quadrant, the cell cultures grown in Ex-Cell Advanced are the least efficient at glycan trafficking through the Golgi apparatus; they are characterized by their large degree of high mannose glycoforms and low degree of the G0F, G1F and G2F glycoforms. Proceeding up to the upper-right quadrant we observed that OptiCHO is more efficient at galactosylation, as evidenced by the decrease in high-mannose glycoforms and increase in the G0F glycoform. However, the still low degree of G1F and G2F glycoforms indicate that cell cultures grown in OptiCHO are still not efficiently achieving terminal galactosylation. The upper-left quadrant contains the cell cultures grown in PowerCHO2 and depict that they are not much more efficient in converting high mannose glycoforms to G0F glycoforms but they are more efficient at converting the G0F glycoforms to G1F and G2F glycoforms. This trend continues into the bottom-left quadrant where cell cultures grown in ProCHO5 exhibit the highest glycosylation efficiency, as characterized by the low degree of high mannose species that are present as well as the conversion of G0F into higher proportions of the terminal galactosylated species.
As the media type is indirectly related to the final glycosylation profile through cellular metabolism, the model results are justified in terms of cell growth indices, nutrient consumption and metabolic byproduct formation over the course of the microbioreactor culture runs. Five daily measurements and 15 parameters derived from the measurements were considered. As the nutrients and metabolic byproducts were measured for 6 days and the cell growth indices were measured for 8 days, there were 141 non-trivial features (the day 0 IVCD parameter is trivial, for example, because it is always 0). We assumed that the variables that differ between cell cultures follow a Pareto distribution; only 20% of the variables account for 80% of the variability seen in the cell culture profiles. Fisher’s F-test was used to eliminate the 80% of the variables that were not useful for discriminating between media types [28]. A PLS model was then built to predict the four glycan categories’ relative abundance from the remaining 29 variables. Variable Importance in Projection was then used to eliminate an additional 16 variables that were not useful for predicting the glycosylation profiles [29]. A final PLS model was built to predict the glycan distribution from the following 13 variables: viable cell density (day 6), growth rate (days 4, 5 and 8), IVCD (day 7), glucose concentration (days 0 and 1), specific glutamine uptake rate (day 2), specific glutamine uptake (on days 2 and 3), total specific glutamine uptake (by days 3-5). The model summary statistics are given in Table 2. The model captures more than 70% of the variability seen in the process measurements and more than 80% of the variability in the glycosylation profiles. Furthermore, the high Q2 value indicates that the results should be reproducible.
| A | R2(X) | R2(Y) | Q2 |
|---|---|---|---|
| 2 | 0.713 | 0.812 | 0.731 |
Table 2: Glycosylation Profiles’ Model’s Summary Statistics
Figure 5 shows the score space and loadings plot for the new PLS model. As the score space appears to follow the same trajectory as the one in Figure 4, it follows that the in-process parameters can be used to explain the glycosylation efficiency trend seen in both score spaces. As cell cultures grown in Ex-Cell Advanced were the least efficient at minimizing high mannose glycoforms (as measured by the (G0F+G1F+G2F)/HM score), their projection was compared against the projections of the other 3 media types together. In doing so, the model coefficients indicate that factors related to glutamine consumption were the most relevant for minimizing high mannose forms. Specifically, lower specific glutamine uptake rate on day 2, a lower amount of glutamine consumed per cell on days 2 and 3 and lower amount of cumulative glutamine consumed per cell on days 3, 4 and 5 were all correlated with a higher degree of G0F, G1F and G2F glycoforms. These findings are summarized in Figure 6.
Figure 5: Partial Least Squares regression model to predict the glycosylation profile from in-process measurements The color coding of the data points in “Loadings” indicate the progression of the microbioreactor run, starting at black for inoculation through tan-colored for harvest, with a gradient of colors in between. The blue data points depict the glycoform loadings. The model coefficients, a product of the glycoform and in-process parameter loadings, indicates that glutamine uptake is correlated with high mannose glycoforms and that modest cell growth and an extended stationary phase is correlated with terminal galactosylation.
Figure 6: Significant Process Parameters and their effects on the abundance of high mannose glycoforms and terminal glycosylation The color coding indicates if the bioreactor process parameters listed on the left are correlated, inversely correlated or not significantly correlated with the two glycosylation outcomes: high mannose forms or terminal glycosylation. Green indicates a positive correlation, red indicates a negative correlation, while grey indicates no significant correlation found.
Since ProCHO5 was the best at maximizing terminal galactosylation (as shown by the (G1F+G2F)/G0F scores), we then evaluated ProCHO5 against OptiCHO and PowerCHO2. Using this comparison, we observed that higher glucose concentrations on days 0 and 1, lower growth rates on days 4 and 5, a lower VCD on day 6, higher growth rates on day 8 and a lower IVCD on day 7 were all correlated with maximizing terminal galactosylation. Evaluating the growth-related parameters together, this is evidence that a more modest degree of cell growth during the exponential growth phase followed by an extended lag phase is the most important factor for promoting a terminal galactosylation. The high glucose concentration correlations may be a spurious relationship, as only the inoculation and day 1 measurements were significant factors for predicting the glycosylation profiles, and it is likely that there are other unmeasured parameters at different levels in the media that are aliased with the glucose concentration that could be preventing glycan processing past G0F. Further evidence of this is the lack of any cell specific glucose parameters or any glucose parameters from later in the cell culture being significant.
Using microbioreactors and different media types we determined that the glycan outcomes of mAb production strongly depend on the nutrients available during the growth process. We initially determined that the different media types demonstrated vastly different terminal galactosylation rates (as measured by (G1F+G2F)/G0F) and formation of high mannose glycoforms (as measured by (G0F+G1F+G2F)/HM). We discovered that high mannose outcomes were not necessarily linked to aglycosylation, which is surprising since the poor nutrient conditions typically responsible for high mannose forms might have also been assumed to result in aglycosylation. To further understand what parameters might be causing these glycan outcomes, we utilized MVDA to establish the in-process variables that best explained our findings. We determined that the factors linked to glutamine consumption were the most important in predicting high mannose outcomes. Specifically, cell culture conditions where glutamine is not heavily consumed will result in glycoforms that are more mature, such as G0F instead of M5. Additionally, early glucose concentrations were positively correlated to terminal galactosylation rates. As both glutamine and glucose are precusors for synthesizing UDP-GlcNAc which is the donor for many different glucosaminyltransferases, it is possible that their influence as precursors is the cause of their effects on the final glycan profile [30]. Factors related to the cell growth rate curves were important in determining the level of terminal galactosylation of the mAbs. Our work reveals important factors in the media that have significant effects on the glycan profile of the produced mAbs, potentially allowing alteration of the media culture conditions in real-time to correct for glycoform profile abnormalities.
Tongzhong Zu and Xiaoshi Wang of the FDA helped with reviewing the manuscript. Partial internal funding and support for this work was provided by the CDER Critical Path Program (CA #1-13). This project was supported in part by the Internship/Research Participation Program at the Office of Biotechnology Products, U.S. Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA.