Journal of Drug Metabolism & Toxicology
Open Access
ISSN: 2157-7609
ISSN: 2157-7609
Research Article - (2018) Volume 9, Issue 2
PH46A is the lead of a new class of bioactive indanes with potential for the treatment of inflammatory bowel disease. A qualitative in vitro metabolism profile of PH46A was investigated in preclinical studies, and the rate of its metabolism in cryopreserved hepatocytes prepared from male Sprague Dawley rat, Beagle dog, Cynomolgus monkey and pooled mixed gender human was compared by LC-MS. The clearance order of PH46A was determined to be rat>dog>monkey>human. The species tested which exhibited the closest clearance values to the human was monkey. Following incubation of PH46A with cryopreserved hepatocytes, 5 metabolites were identified, including M1 (keto-PH46), M2 (PH46-OH, PH132), M3 (PH46-diOH), M4 (keto-glucuronide-PH46) and M5 (glucuronide conjugate-PH46). It was found that the human metabolites M2 and M5 were also present in rat, dog and monkey, while M1 was present in all species except monkey. M2 was detected in dog and monkey by LC1 conditions, but only in dog by LC2. Therefore, the metabolism in rat was most similar to human, in terms of the metabolites observed, but all putative human metabolites were present in rat and dog. We further explored the characterization of key metabolite M2 (PH46-OH). Identical PH46-OH was obtained via a bio-catalytic oxidation method from PH46 using rat liver microsomes (RLM) and the human liver P450s (Cyp2D6, Cyp 2C19 and Cyp 4A11) following screening of selectAZyme panels of microbial P450s, recombinant human liver P450s and different microsomes. RLM was used in scale up production and PH132 was isolated and characterized as 4-(((1’S,2’S)-1’,6-dihydroxy-1’,3’-dihydro1H,2’H-[2,2’-biinden]-2’-yl)methyl) benzoic acid via LC-MS/MS, NMR and HRMS. The site of hydroxylation on the biindane scaffold was unexpected. The outcomes of these studies have provided valuable information for future pharmacokinetic and in vivo toxicological investigations.
Keywords: Bioactive indane; PH46A; PH46; Metabolism; PH132; LC-MS; Biosynthesis
A wide range of bioactive indane based scaffolds are employed clinically as treatment of many disease states, including inflammatory [1-3], cancer [4,5] and neurological conditions [6] and also in the treatment of HIV [7]. Several classes of diastereoisomeric and enantiomeric molecules containing the indane moiety have been developed and characterized by our group [8-10]. Smooth muscle relaxation and mediator release inhibition properties have been demonstrated for many of these novel compounds [11-13]. In continuation of our work, we more recently designed and synthesized a series of dimeric indane compounds, with two contiguous stereogenic centres, one of which is a quaternary centre. Some of these compounds possess potent anti-inflammatory activity, in particular, a lead chiral molecule PH46A with S,S configurations [14-18], has been identified by our group and was subject to a range of preclinical studies prior to entering a Phase I clinical trial [19].
ADME (Absorption, distribution, metabolism, and excretion) is one of the major elements involved in the drug discovery and development process. In the last two decades, comparative in vitro metabolism studies of different species has been widely employed as a more costeffective approach to understand the animal toxicological results and the relevance to humans, and to characterize new drug candidates in the early drug discovery phase [20]. In the present study, we conducted an in vitro metabolism study of PH46A in cryopreserved hepatocytes of male Sprague Dawley rat, Beagle dog, Cynomolgus monkey and pooled mixed gender human in order to: (a) to identify the potential metabolites of PH46A and pathways of metabolism in each species; (b) to identify the presence/absence of unique human metabolites; (c) to compare the rates of metabolism of PH46A by determination of loss of PH46A across the species using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Based on the metabolism data obtained, the major metabolite across all species, M2 hydroxylated PH46 (PH46-OH or PH132) was further studied in order to support the next phase of pharmacokinetic investigation. The work was started with an extensive screening of 18 recombinant human liver P450 monooxygenases, 40 microbial P450 monooxygenases and 4 microsomes against PH46, followed by LC-MS/ MS analysis with the aim of identifying the metabolite PH132 observed in the metabolism study. The cytochrome P450 monooxygenases (P450s or Cyps) are a large family of heme protein containing enzymes found in animals, plants and microorganisms, which are involved in drug metabolism introducing a single oxygen atom derived from molecular oxygen into an organic molecule [21,22]. They catalyse a variety of reactions, such as hydroxylation of aliphatic and aromatic carbons, oxidation of organic nitrogen and sulphur, epoxidation and Baeyer-Villiger oxidation. Liver microsomes (LM) contain a cocktail of different cytochromes P450, flavin-monooxygenases, carboxylesterases, epoxide hydrolases, UDP-glucuronosyltransferases and other drugmetabolizing enzymes and therefore have a high success rate for any desired oxidation reaction [23]. In this paper, we also report the scaleup production via the chosen biotransformation system, isolation and characterization of this key metabolite PH132. The potential chirality change of PH46 (S,S) with formation of the diastereomer to PH46, (R,S)-PH46, was also monitored.
Since PH46A salt dissociates in solution, the measurements of PH46A by LC-MS analysis throughout the two studies are related to the parent PH46 (free acid form). Based on structural similarities, compound 1 was chosen as internal standard (IS) in the metabolism study (Figure 1).
Chemicals
Test item PH46A, reference compounds [PH46, diastereomer (R,S)-PH46 & keto-PH46] and Compound 1 (IS) were obtained inhouse and used after identity/purity checking. Dulbecco’s Modified Eagle Medium (DMEM) was purchased from Invitrogen (UK). Tetrabutylammonium phosphate monobasic was purchased from Sigma-Aldrich (UK). 0.9% saline was obtained from Baxter (UK). All solvents (as anhydrous, HPLC or general process grades) were obtained either commercially or locally.
Biological reagents
Male Sprague Dawley rat cryopreserved hepatocytes (Lot No. RJW), male Beagle dog cryopreserved hepatocytes (Lot No. KLI and HTU), Male Cynomologus monkey cryopreserved hepatocytes (Lot No. YHC), and mixed gender human cryopreserved hepatocytes (Lot No. NQC) were obtained from Celsis, In vitro Technologies (MD, USA). All preparations were stored in liquid nitrogen prior to use. [14C]-Testosterone (Batch No. 3590023), testosterone (Batch No. 070M1626V) and [14C]-7-ethoxycoumarin (7-EC) (Batch No. RUS1277) were purchased from Perkin Elmer (UK), Sigma-Aldrich (UK) and Cypex (UK), respectively. All select AZymeTM enzymes were obtained from Almac (UK) as freeze dried cell free exacts.
Animals and husbandry
Six male Sprague Dawley rats, aged 10-12 weeks at dosing, were supplied by Charles River (UK) and surgically prepared with an indwelling femoral vein cannula. After a post-surgery recovery of ca 7 days and prior to dose administration, a pre-study health evaluation was conducted on these animals and the results found to be satisfactory. A standard laboratory diet and mains tap water was available to the animals. Clinical signs were monitored throughout the study in order to asses any reaction to treatment.
Incubation conditions for cryopreserved hepatocytes
The primary stock solution of PH46A (4 mM) was prepared in distilled water, which was further diluted using distilled water to give the secondary stock solution (1 mM). PH46A dosing solutions were prepared at 40 and 10 μM in culture medium DMEM from the stock solutions.
A single pooled preparation from each species was used. Hepatocytes from each species (rat, dog, monkey and human) were thawed according to the suppliers’ instructions. The hepatocyte viability of each preparation was determined using the trypan blue exclusion assessment by calculating the percentage of live cells present [24]. Incubations were performed, in triplicate, in 12 well plates in an incubator with temperature of 37ºC in a humidified atmosphere of 95% air & 5% CO2. Test reactions contained PH46A (5 μM or 20 μM in water) and hepatocytes (1 x 106 cells/mL) in a total volume 1 mL of (DMEM). Each metabolic reaction was terminated after 0, 30, 60, 90, 120 and 240 min by removing an aliquot (100 μL) from the appropriate well at the appropriate time point and transferring to 100 μL of cold acidified MeOH containing IS (5 μM). After the last timepoint sampling, the remaining sample (400 μL) was mixed with cold acidified MeOH (400 μL) containing IS. All samples were stored at -20ºC until being processed. Prior to LC-MS analysis, the samples were thawed and the aqueous MeOH supernatants were obtained following centrifugation (10,000 rpm for 10 min). Control incubations were also performed in the absence of hepatocytes to test the stability of PH46A (5 μM).
For the metabolite identification phase, PH46A samples (20 μM in water) were incubated for 240 min only in DMEM (1 mL) with sampling at 0 and 240 min and terminated by an equal volume of cold acidified MeOH containing IS.
The metabolic competence of the cells (1 x 106 cells/mL) was assessed in parallel following incubations of [14C]-7-EC (10 μM in DMEM) and [14C]-testosterone (150 μM in ACN) in DMEM (1 mL) as positive control. Aliquots (100 μL) were taken and terminated at different time points and the samples were prepared for analysis as above. The remaining samples (400 μL) were mixed with the appropriate termination reagent.
Analytical methods validation & determination of PH46A in culture medium
An LC-MS/MS method for detection and quantification of PH46 in dosing solutions and post-incubation supernatants was developed, and the following parameters (assay linearity, intra-batch assay accuracy and precision) were assessed.
Assay linearity: A series of calibration standards (CS) were prepared in DMEM containing PH46 over the target concentration range of 0.5-7.0 μM at a minimum of 6 concentrations, and a fixed concentration of IS. Calibration curves were constructed from PH46: IS peak area response ratios plotted against the actual concentration of PH46 to determine the optimum regression parameters. The acceptance criteria was that the calculated concentration of PH46 for each standard from the calibration line should be within 100 ± 15% of the actual amount in culture media [100 ± 20% at lower limit of quantification (LLOQ)]. At least 75% (and a minimum of 6) of the CS samples should meet the above criteria.
Intra-batch assay accuracy and precision: Replicate (n=6) quality control (QC) samples were prepared in DMEM containing PH46 at 0.5, 1.2, 2.6 and 5.6 μM and a fixed concentration of IS. The samples were extracted and analyzed, and the concentration of each replicate sample was determined from the calibration curve made above. The acceptance criteria at each level were that the intra-batch assay accuracy (the mean percentage determined concentration/actual concentration) should be within 100 ± 15% (100 ± 20% at LLOQ) and the intra-batch assay precision (the coefficient of variation of the mean determined concentration) should be ≤ 15% at each concentration level (≤ 20% at LLOQ).
Chromatographic conditions: The culture medium samples were analyzed by LC-MS/MS and the concentration levels of PH46 in the samples were determined using IS. The LC-MS analysis was performed on Perkin Elmer series 200 and AB Sciex API 3000. Ascentis Express C18 column (75 x 2.1 mm, 2.7 μm) with guard column (PreFrit Filter, 6.4 x 1.6 mm, 0.5 μm) was utilized with mobile phases: A [MeOH/acetic acid (AA), 100/0.2, v/v] and B (water/AA, 100/0.2, v/v). The pump gradient (A/B) was: 0 min (60/40), 2 min (80/20), 4 min (80/20), 4.5 min (100/0), 4.7 min (60/40), 4.9 min (100/0) and 8.0 min (60/40) with flow rate of 0.3 mL/min. The column and autosampler temperatures were 60°C and 4ºC respectively. The injection volume was 1-20 μL (as required for sensitivity) with run time of 10 min (taking into account autosampler cycle time)]. The mass spectrometer parameters were Turbo Ion Spray ionisation negative mode, ion spray temperature of 550°C, probe position of 3 mm (X) & 7 mm (Y), standard API3000 all nitrogen gases, settling time of 5 msec and pause time of 5 msec.
Calculations & run acceptability: Integration of PH46 and IS peaks was done by a smoothing factor of 2 and the peak area ratio of PH46:IS for each CS, QC and test samples were calculated. A calibration curve was constructed by plotting the peak area ratio of the calibrations standards vs. the concentration of each test item in matrix and determine the linear regression parameters of the curve, using a weighting factor of 1/x2 (1/concentration2). The concentration of PH46 in QC samples and test samples was determined by interpolation of the peak area ratios from the calibration curve. At least 75% of CS samples must back-calculate to within 100 ± 15% of the actual concentrations (100 ± 20% at the lower limit of quantification). At least 66% of QC samples must be within 100 ± 15% of the actual concentration, including at least one at each level.
Each batch contained double blank sample (absence of both PH46A and IS), single blank sample (absence of PH46A and presence of IS), CS samples, QC samples and test samples. The CS samples were prepared at 0.5, 1.3, 1.8, 2.3, 2.7, 3.2, 3.7, 4.5, 5.0, 6.2 and 7.0 μM, which were injected at the start of each batch. Duplicate LLOQ (0.5 μM) and upper limit of quantification (ULOQ) (7.0 μM) was also made to minimize the risk of not achieving a valid standards at LLOQ or ULOQ levels. Only one of the CS samples, at each concentration level, was employed in the assay calibration line. QC samples were prepared in duplicate at least at 1.2, 2.6 and 5.6 μM. For the analysis of test samples groups of one at each of low, medium and high concentrations of the QC samples were analyzed after the calibration line and after study samples. Where QC samples are prepared in triplicate, additional groups of QC samples should be interspersed within the test samples. Study samples were run bracketed by groups of QC samples. The test samples were analyzed from low to high concentrations. Under normal assay conditions two reconstitution solvent samples MeOH/water/AA (50/50/0.2, v/v/v) samples were used after the top standard and high QC samples to avoid significant carryover.
HPLC analysis of supernatants from [14C]-7-EC & [14C]-testosterone incubations
[14C]-7-EC, [14C]-Testosterone and their corresponding metabolites present in the post-incubation supernatants were resolved by HPLC analysis performed on Waters 2695 separations module with Waters 486 Tunable absorbance detectors and Perkin Elmer Radiomatic™ Flo-one ®\Beta Scintillation Analyser (Model 150TR). For [14C]-7-EC incubation, Hypersil BDS C8 column (250 x 4.6 mm, 5 μm) was used with mobile phases: A [tetrabutylammonium phosphate monobasic in water (5 mM)/AA, 100/1, v/v] and B (ACN/AA, 100/1, v/v). The pump gradient (A/B) was: 0 min (94/6), 10 min (49/51), 20 min (49/51), 25 min (94/6) and 35 min (94/6) with flow rate of 1 mL/min. The column and auto sampler temperatures were ambient and 10°C, respectively. The injection volume was 0.1 mL with run time of 35 min. UV wavelength 320 nm was used with data captured by Labsystems Atlas 2002 R1. For [14C]-testosterone incubations, Novapak C18 column (150 x 3.9 mm, 4 μm) and pre-Column Novapak C18 (20 x 3.9 mm, 4 μm) were used with mobile phases: A (water), B (MeOH) and C (ACN). The pump gradient (A/B/C) was: 0 min (75/25/0), 40 min (30/64/6), 45 min (75/25/0) and 55 min (75/25/0), with flow rate of 1 mL/min. The column and autosampler temperatures were 50°C and 10°C, respectively. The injection volume was 0.1 mL with run time of 55 min. UV wavelength of 240 nm was used with data captured by Labsystems Atlas 2002 R1.
Calculation of intrinsic clearance
Intrinsic clearance values were calculated using the following equation. The physiological parameters (no. of hepatocytes per g of liver & g of liver per kg of bodyweight) used were: 120 x 106 & 40 for Rat [25], 240 x 106 [26] & 32 [27] for dog, 135 x 106 & 25 for monkey [28] and 120 x 106 & 26 for human [25].
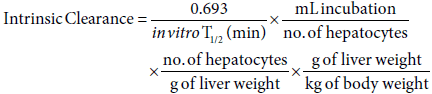
Where in vitro T1/2 is estimated using the gradient (k) of the line of the logarithmic mean % parent compound remaining against, and the equation: in vitro T1/2 = 0.693/-k.
Analytical method for metabolite identification
The metabolite profiling investigation following incubation was performed by LC-MS/MS analysis using Shimadzu LCMS-IT-TOF mass spectrometer and Shimadzu Prominence HPLC with pumps (LC- 20ADXR), degasser (DGU-20A3), autosampler (SIL-20ACXR), column oven (CTO-20ACXR), DAD (SPDM20AXR) and communications module (CBM-20AXR).
LC conditions 1 (LC1): Agilent EC-C18 column (50 × 2.1 mm, 1.7 μm) was used with mobile phases: A (water/AA, 100/0.2, v/v) and B (MeOH/AA, 100/0.2, v/v). The pump gradient (A/B) was: 0 min (70/30); 11 min (0/100); 12 min (0/100); 13 min (70/30) and 15 min (70/30) with flow rate of 0.3 mL/min. The column and autosampler temperatures were 60°C and 4°C, respectively with diode array detector (DAD) range of 190-800 nm.
LC conditions 2 (LC2): Kinetix PFP column (50 × 2.1 mm, 1.7 μm) was used with mobile phases: A (water/AA, 100/0.2, v/v) and B (MeOH/ AA, 100/0.2, v/v). The pump gradient (A/B) was: 0 min (90/10); 11 min (0/100); 12 min (0/100); 13 min (90/10) and 15 min (90/10) with flow rate of 0.3 mL/min. The column and autosampler temperatures were 60°C and 4°C with DAD range of 190-800 nm.
MS conditions: Electrospray ionisation (ESI) was selected with both curved desolvation line and block temperatures at 230°C. The nebulising gas (N2) flow was 1.5 L/min with collision-induced dissociation gas of Argon and drying gas on. The data acquisition in both positive and negative mode was achieved using automatic gain control (ASC 10e6) and the Q (low mass cut-off) value was 0.251 (45 kHz) with precursor ion isolation of 3 amu.
Accurate mass measurement was used to differentiate between potential metabolites with the same nominal mass. The accurate mass error (ppm) was calculated from a comparison of the measured mass with the theoretical mass for a suggested assignment. A low accurate mass error (< 5 ppm) supports the assignment. Conversely a large accurate mass error would refute the assignment. Metabolite screening software (Met ID Solutions version 1.2) was also used to aid the identification of potential metabolites. Analysis of T0 sample was used to subtract background matrix for each species. A media control sample was used to indicate any non-metabolic changes to PH46. The output of the software screening was reviewed and possible metabolites investigated further manually.
Screening of enzymes & microsomes against PH46 & LC-MS detection
Almac’s selectAZyme panels of 18 recombinant human liver P450 monooxygenases, 40 recombinant microbial P450 monooxygenases (self-sufficient cytochrome P450 enzymes were heterologously expressed in E. coli) and 4 different microsomes were screened against PH46 using a glucose dehydrogenase based cofactor regeneration system. Reactions were analyzed after 4 h and overnight reaction time by newly developed LC-MS method, LC3, in both positive & negative modes, and the tentative metabolite hits were further analyzed via MS/MS experiments. The details of the screening procedure and the corresponding tentative metabolite screening results are provided in Supplementary Table S1.
LC conditions 3 (LC3) (for screening purpose only): Luna C18 (50 x 2 mm, 3 μm) was used with mobile phases: A (water/AA, 100/0.1, v/v) and B (ACN/AA, 100/0.1, v/v). The pump gradient (A/B) was: 0 min (70/30), 5 min (35/65), 5.10 min (0/100), 7 min (0/100), 7.10 min (70/30) and 10 min (70/30) with flow rate of 1 mL/min. The column temperature was ambient temperature.
LC conditions 4 (LC4): It was further developed for all further analysis throughout the biotranformation study, due to the most accurate match of the relative retention times of the metabolites observed in the metabolism study. Kinetix C18 (50 x 4.6 mm, 2.6 μm) was used with mobile phase: A (water/AA, 100/0.2, v/v) and B (MeOH/ AA, 100/0.2, v/v). The pump gradient (A/B) was 0 min (70/30), 11 min (0/100), 12 min (0/100), 13 min (70/30) and 15 min (70/30) with flow rate of 0.5 mL/min. The column temperature was 60°C.
MS conditions: AB Sciex QQQ 365; Scan type: Q1 MS; polarity: positive/negative; ion source: turbo spray; mass range: 100-700; NEB: 8; CUR: 8; IS: 4200; TEM: 300; DP: 46; FP: 180; EP: 9.
Scale-up production of PH132 via selected bio-oxidation system and characterization methods
The procedure outlined below was performed on 500 mL scale and total of 12 x 500 mL reactions were conducted. KH2PO4 buffer (100 mM, 500 mL) were prepared containing MgCl2 (5 mM), with pH adjustment to 7.4 using KOH solution. 437.5 mL of such buffer was then transferred into 2 L Erlenmeyer flask, followed by additions of Glucose (3.6 g) and GDH (2 g). The resulting mixture was gently stirred until complete dissolution, followed by the addition of 1 mL of PH46 stock solution (262 mM in DMSO) and 12.5 mL of RLM. The reaction was started with the addition of nicotinamide adenine dinucleotide phosphate Na-salt solution (40 mM, 50 mL) and was left shaken overnight at 37ºC at 200 rpm. The flasks were “closed” with a punctured tin foil to allow for oxygen transfer. ACN (125 mL) was added to quench each reaction, and the resulting solids (from proteins precipitation) were removed by centrifugation at 8000 rpm for 10 min. The residual solution was analyzed by LC-MS method using LC4 to determine conversions and subsequently prepared for sold phase extraction (SPE) and prep-HPLC purification.
SPE: A 60 mL Strata-X (33 u, polymeric reverse phase) tube was initially washed with MeOH (150 mL) and equilibrated with water (300 mL), followed by sample loading (1500 mL of three reactions for the column). The flow through and the water washes were collected and analyzed for the desired product by HPLC. After sample loading, the tube was subsequently washed with water (300 mL). Before the tube became dry, it was wetted with MeOH. MeOH was incubated on the column for 5 min without flow. The material was subsequently eluted with MeOH (150 mL) at minimum flow. More MeOH was used for the complete elution, which was checked by HPLC. All fractions were combined and diluted to a 20% MeOH solution with water for the next step, prep-HPLC purification.
Prep-HPLC purification was performed on Luna semi prep-HPLC column (250 x 30 mm, 10 μm) with 750 mL of sample (containing 300 mg of crude material after dilution) applied to the column via an external pump operating at 9 mL/min. The isocratic mobile phase system, ACN/Water/AA (40/60/0.1, v/v/v), was used with flow rate of 42.5 mL/min for each injection (600 mg of crude were created in total 2 injections). The total run-time was 70 min. The purity of the fractions was then tested by LC4 and the fractions containing PH132 with purities >95% were pooled and concentrated to dryness by vacuo at 37ºC. ACN was used to azeotroph any residual water. The fractions with less purity were combined and purified further.
NMR spectra were recorded on Bruker DPX-600 and Bruker DPX- 400 instruments operating at 400.1 MHz and 600.1 MHz for 1H NMR; 100.6 MHz and 150.9 MHz for 13C-NMR and calibrated using residual undertreated solvent as an internal reference (CHCl3@ δ 7.26 ppm 1H NMR, δ=77.16 13C NMR; DMSO-d6@ δ=5.32 ppm 1H NMR, δ=39.51 ppm 13C NMR). The following abbreviations were used to explain 1H NMR multiplicities: s=singlet, d=doublet, t=triplet, m=multiplet, br=broad. NMR spectra were analyzed with Bruker Toppsin 3.5pl7 software. High-resolution mass spectra (HRMS) were measured on a Micromass LCT electrospray TOF instrument with a WATERS 2690 autosampler and methanol as carrier solvent. Data analysis was performed using MassKynx software.
Assay linearity, intra-batch assay accuracy and precision
It was found that regression analysis of the peak area ratios of PH46:IS against the amount of PH46 had demonstrated good linearity in culture medium over the range 0.5-7 μM with a 1/x2 linear weighting. The determined concentrations for these standards (0.50, 1.25, 1.75, 2.25, 2.69, 3.23, 3.74, 4.49, 4.99, 6.24 and 6.98 μM) met the acceptance criteria. The intra-batch accuracy and precision of the assay for PH46A in DMEM met the acceptance criteria for each concentration level (0.50, 1.24, 2.64 and 5.60 μM) with replicate (n=6). The actual results are giving in Supplementary Tables S2 and S3.
Incubation of PH46A (5 μM) with cryopreserved hepatocytes
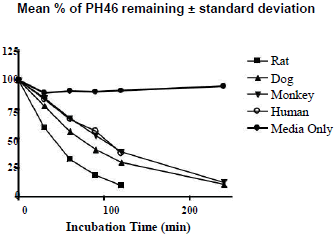
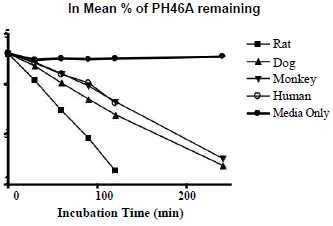
The hepatocyte viability assessment for all species was above 75% acceptance criterion with the viability values of 84.3% (male rat), 80.7% (male dog), 78.0% (male monkey), 76.4% (mixed gender pooled human). Incubation of PH46A at 5 μM with cryopreserved hepatocytes resulted in partial loss of PH46 in all four species at all-time points, except T=0 min, over the time course of the incubation of 240 min. This loss was not observed significantly during incubation of PH46A with DMEM, indicating that PH46 appeared to be stable over the time course under incubation conditions; however small portion (~10%) of PH46 loss was seen after 30 min of incubation in DEME, which was considered to be due to weak interaction between PH46 and amino acids present in DEME media.
The concentration of PH46 remaining at 240 min was found to be below LLOQ (0.5 μM) following incubation with rat and human cryopreserved hepatocytes. The percentage of PH46 remaining at 240 min was 10.8% and 12.4% following incubation with dog and monkey hepatocytes, respectively. The results are presented in Table 1. The natural log (ln) of the percentage of PH46 remaining was also calculated. The in vitro intrinsic clearance values following incubation at 5 μM of PH46A (mL/min/kg body weight) were also determined to be 93.1 (rat), 72.2 (dog), 30.0 (monkey) and 24.4 (human). The order of clearance for the species was rat>dog>monkey>human.The concentration of PH46 remaining at 240 min was found to be below LLOQ (0.5 μM) following incubation with rat and human cryopreserved hepatocytes. The percentage of PH46 remaining at 240 min was 10.8% and 12.4% following incubation with dog and monkey hepatocytes, respectively. The results are presented in Table 1. The natural log (ln) of the percentage of PH46 remaining was also calculated. The in vitro intrinsic clearance values following incubation at 5 μM of PH46A (mL/min/kg body weight) were also determined to be 93.1 (rat), 72.2 (dog), 30.0 (monkey) and 24.4 (human). The order of clearance for the species was rat>dog>monkey>human.
 |
 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rat | Dog | Monkey | Human | DMEM Only | Rat | Dog | Monkey | Human | DMEM Only | |
| 0 | 100 ± 6.55 | 100 ± 5.69 | 100 ± 6.89 | 100 ± 2.99 | 100 ± 3.81 | 4.61 | 4.61 | 4.61 | 4.61 | 4.61 |
| 30 | 59.3 ± 0.32 | 77.9 ± 1.72 | 84.7 ± 1.62 | 83.8 ± 3.72 | 88.9 ± 1.19 | 4.08 | 4.36 | 4.44 | 4.43 | 4.49 |
| 60 | 32.3 ± 0.94 | 56.1 ± 0.69 | 67.3 ± 4.82 | 66.6 ± 2.25 | 90.6 ± 1.18 | 3.48 | 4.03 | 4.21 | 4.20 | 4.51 |
| 90 | 18.5 ± 1.59 | 40.5 ± 1.81 | 52.7 ± 0.78 | 56.3 ± 7.45 | 90.0 ± 0.41 | 2.92 | 3.70 | 3.96 | 4.03 | 4.50 |
| 120 | 9.77* | 29.7 ± 1.34 | 38.4 ± 2.23 | 37.6 ± 1.65 | 90.9 ± 1.07 | 2.28* | 3.39 | 3.65 | 3.63 | 4.51 |
| 240 | 10.8 ± 0.58 | 12.4 ± 1.36 | 94.74 ± 5.26 | 2.38 | 2.52 | 4.55 | ||||
Table 1: Percentage (left) and ln percentage (right) of PH46 remaining following incubation of PH46A with cryopreserved hepatocytes.
Incubation of [14C]-7-EC and [14C]-testosterone with cryopreserved hepatocytes
Incubations of cryopreserved hepatocytes with [14C]-7-EC and [14C]-testosterone resulted in partial or complete disappearance of parent compounds over 240 min incubation periods. In the case of [14C]-7-EC, 3 of the metabolites formed were identified in all species against authentic standards: 7-hydroxycoumarin (7-HC) (phase I), 7-HC glucuronide and 7-HC sulphate (phase II). For [14C]-testosterone, the major metabolite 6β-hydroxytestosterone was identified against an authentic standard in all species. The above data indicates that all hepatocytes were intact and were suitable for the assessment of metabolism of PH46A.
Metabolite profiling of PH46A in rat, dog, monkey and human hepatocytes
Reference standard PH46 was analyzed first. Accurate mass measurement of the deprotonated molecular ion (m/z 381 [M-H]-) with retention time (RT) 7.9 min confirmed the formula C26H22O3. The corresponding protonated ion was not observed, which was believed to be due to loss of water in the mass spectrometer source resulting in the presence of the ion with m/z 365 [M+H]+. Two potential routes were considered as two potential fragment ion structures were possible for the fragment ions with m/z 261 & 245. Fragmentation resulting in the loss of the indanol moiety was supported by accurate mass measurement. A fragmentation pathway was postulated based on analysis of the free acid form (PH46) of PH46A (Figure 2).
The representative chromatograms and mass spectra are given in Supplementary Figure S1. Comparison of the fragment ions observed for PH46 and postulated metabolites was used to support metabolite identification. During the metabolite profiling study, PH46 and metabolites (M1-M5) were detected in the same species by LCMS analysis at 240 min under both LC1 and LC2 conditions, with the exception of M4 (Figure 3). A summary of metabolite detection by LC1 for each species and the corresponding UV chromatograms as representative are presented in Table 2 and Figure 4. The details of each metabolite’s identification are described below and the related chromatograms and spectra of each metabolite are given in Supplementary Figures S1-S5.
| Species | RT (min) | m/z ([M-H]-) | Postulated ID |
|---|---|---|---|
| Rat | 6.445 | 397 | M2 |
| 6.605 | 397 | M2 | |
| 8.032 | 381 | PH46 | |
| 8.556 | 379 | M1 | |
| Dog | 6.439 | 397 | M2 |
| 6.608 | 557 | M5 | |
| 6.874 | N/A | N/A | |
| 7.326 | 555 | M4 | |
| 8.033 | 381 | PH46 | |
| 8.556 | 379 | M1 | |
| Monkey | 6.434 | 397 | M2 |
| 6.605 | 557 | M5 | |
| 8.032 | 381 | PH46 | |
| Human | 6.445 | 397 | M2 |
| 6.608/6.875 | 557 | M5 | |
| 8.032 | 381 | PH46 | |
| 8.556 | 379 | M1 |
Table 2: Summary of metabolite detected by LC-MS/MS (240 min, LC1) for each species (M1:keto-PH46; M2: PH46-OH (PH132); M4: keto-glucuronide-PH46; M5: glucuronide-PH46).
PH46: The deprotonated ion m/z 381 ([M-H]-) was observed at RT of ca. 8.15 min (LC1) which was consistent with PH46. Accurate mass measurement confirmed the molecular formula as C26H22O3, and the product ions m/z 261, 245, 135 and 115 ([M-H]-) observed following fragmentation was consistent with PH46. A response for the positive ion m/z 365 ([M+H]+), formed by loss of water in the mass spectrometer source was also observed. This in-source fragmentation of the positive molecular ion was also consistent with the PH46 reference standard analysis and therefore was deemed to reinforce the identification of PH46 in hepatocyte incubates.
M1 (keto-PH46) with m/z 379 (RT ca. 8.7 min): The ion m/z 379 ([M-H]-) were observed for each species except monkey, which were postulated to correspond to keto-PH46 (C26H20O3) with RT of 8.7 min in the extracted ion chromatogram (EIC), and confirmation was obtained from accurate mass measurement. Fragmentation of the deprotonated molecular ion resulted in the product ions m/z 244 & 135 which was consistent with the fragmentation of the parent PH46 and thus supports the identification. A representative example is shown in Supplementary Figure S2.
M2 (PH46-OH, PH132) with m/z 397 (RT ca. 6.5 min): The ion m/z 397 ([M-H]-) were observed for each species. Accurate mass measurement confirmed the formula C26H22O4 corresponding to PH132. Fragmentation of the deprotonated molecular ion resulted in the product ions m/z 261 and 135. These fragment ions were consistent with PH46 fragmentation and suggested hydroxylation may occur on the indanol moiety (Supplementary Figures S3-S9). The spectra observed at 6.5 and 6.8 min were of low intensity which may be due to suppression by other ions such as m/z 557 which was not related to PH46 metabolism based on accurate mass observed.
M3 (PH46-diOH) with m/z 413 (RT ca. 6.3-7.5 min): The ion m/z 413 ([M-H]-) were observed in rat and the presence of peaks with RTs ca. 6.3-7.5 min in the EIC using LC2 indicated several isomers could be possible (Figure S5). Accurate mass measurement of the molecular ion confirmed the formula C26H22O5 corresponding to PH46-diOH. Fragmentation of the deprotonated molecular ion resulted in the product ions m/z 395 & 377.
These fragments suggested the consecutive loss of two molecules of water from PH46-diOH and it was postulated that hydroxylation may occur on adjacent positions within a ring moiety. A structure for the fragment ion m/z 395 has been suggested but a mechanism by which a second loss of water from the ion m/z 395 to yield ion m/z 377 was not clear. It is envisaged that dihydroxylation could be possible on any of the 3 ring moieties, but it was not possible to confirm this from the product ions obtained by current study.
M4 (keto-glucuronide-PH46) with m/z 555 (RTs ca. 8.0-8.2 min): The ion m/z 555 ([M-H]-) were observed for monkey and dog at 240 min by LC1 and for dog only by LC2, which were postulated to correspond to a ketone glucuronide of PH46. The presence of several peaks with RTs ca. 8.0-8.2 min in the EIC by LC2 indicates that several isomers (as with the glucuronide) are possible (Figure S6). Accurate mass measurement confirmed the formula C32H28O9. The deprotonated molecular ion fragmentation resulted in the product ion m/z 379 which were in turn fragmented resulting in product ions m/z 245 & 135, consistent with those formed following PH46 fragmentation. The spectra observed ca 8.0-8.2 min were of low intensity which may be due to suppression by other ions such as m/z 521.
M5 (glucuronide conjugate-PH46) with m/z 557 (RT ca. 7.3-7.7 min): The ion m/z 557 ([M-H]-) were observed for each species using both LC1 & LC2. An example of rat sample analysis via LC2 is given in Figure S7. These were postulated to correspond to glucuronide- PH46 and several unresolved peaks with RT ca. 7.3-7.7 min in the EIC indicated that several isomers were possible (Figure S8). These could be postulated to be the result of acyl migration and the analysis using LC1 afforded better resolution of the possible isomers (Figure S9). Accurate mass confirmed molecular formula C32H30O9 corresponding to glucuronide-PH46. Fragmentation of the deprotonated molecular ion resulted in ion m/z 381, which was further fragmented forming product ions consistent with those formed following fragmentation of PH46.
The results showed that PH46 was observed in all samples at 0 and 240 min and PH46A with hepatocytes resulted in the formations of 5 metabolites among various species: M1 (keto-PH46), M2 (PH46- OH, PH132), M3 (PH46-diOH), M4 (keto-glucuronide-PH46), M5 (glucuronide conjugate-PH46). LC-MS experiments via both LC1 and LC2 conditions provided the prominent and informative product ions for structural elucidation Table 3 and a summary of the qualitative metabolite formation following incubation is shown in Table 4. The postulated fragmentation pathways of all 5 metabolites are given in Figure 5.
| Component | RT (min) | Nominal mass | Formula | Characteristic ions |
|---|---|---|---|---|
| LC1/LC2 | (Accurate mass) | |||
| [M-H]- | ||||
| PH46 | 8.0 / 8.7 | 381 (381.1496) | C26H22O3 | 261, 245, 135, 115 |
| M1 | 8.7/ 9.2 | 379 (379.1339) | C26H20O3 | 244, 135 |
| M2 | 6.5/ 7.7 | 397 (397.1445) | C26H22O4 | 261, 135 |
| M3 | 3.8/ 6.3 | 413 (413.1394) | C26H22O5 | 395, 377 |
| M4 | 7.4/ 8.0 | 555 (555.1661) | C32H28O9 | 379, 245, 135 |
| M5 | 6.7/ 7.5 | 557 (557.1817) | C32H30O9 | 381, 261, 245 |
Table 3: Product ions of PH46 and its 5 metabolites observed by LC-MS. M1: keto-PH46; M2: PH46-OH; M3: PH46-diOH; M4: keto-glucuronide-PH46; M5: glucuronide conjugate-PH46.
| LC conditions | Species | PH46 | M1 | M2 | M3 | M4 | M5 |
|---|---|---|---|---|---|---|---|
| LC1 | PH46 reference | √ | - | - | - | - | - |
| Rat | √ | √ | √ | √ | - | √ | |
| Dog | √ | √ | √ | - | √ | √ | |
| Monkey | √ | - | √ | - | √ | √ | |
| Human | √ | √ | √ | - | - | √ | |
| DMEM | √ | - | - | - | - | - | |
| LC2 | PH46 reference | √ | - | - | - | - | - |
| Rat | √ | √ | √ | √ | - | √ | |
| Dog | √ | √ | √ | - | √ | √ | |
| Monkey | √ | - | √ | - | - | √ | |
| Human | √ | √ | √ | - | - | √ | |
| DMEM | √ | - | - | - | - | - |
Table 4: Summary of metabolite detection by LC-MS (240 min, LC1 & LC2) for all species.
The metabolic pathways shared similarities in all species tested. Two common metabolic routes to all species appeared to be phase I hydroxylation and phase II glucuronidation resulting in the formations of M2 and M5, respectively. In addition, oxidative transformation as minor metabolic pathway was detected in all species except monkey samples due to absence of M1 under both LC1 and LC2 conditions. M3 as unique metabolite was witnessed in rat hepatocyte sample. Furthermore, the observation of M4 in dog and monkey samples suggested that PH46 was susceptible to oxidation at hydroxyl functionality, followed by glucuronidation conjugation reaction. However, M4 was not detected in monkey samples under LC2. It is important to note that no major human-specific metabolite was found. Due to the qualitative nature of the current work, it was not possible to provide quantification of the metabolites detected. The relative quantification between T0 and the T240 in each species may be inferred from the UV chromatograms; however, the comparison of UV chromatograms was required with caution.
Recombinant enzymes and microsomes screening
The proposed route to the generation of the desired PH132 metabolite by biocatalysis involved initial screening using microbial P450s, recombinant human liver P450s and microsomes, tentative hits were observed after 4 h with rat liver microsomes (RLM) (~7% conversion) and human Cyp-3 (Cyp2D6, ~11% conversion), human Cyp-4 (Cyp2C19, ~11% conversion) and human Cyp-13 (Cyp4A11, ~25% conversion). The result of the RLM reaction after 24 h incubation via LC3 showed that the hydroxyl group in PH46 was unstable under the positive mode, with observations of 365 amu (PH46-H2O) and 381 amu (PH132-H2O) only (see chromatograms and spectra in Figure 6).
During the MS/MS experiments of the RLM sample against reference PH46, fragmentation patterns in both negative mode (135 & 261 amu) and positive mode (131, 135, 246 & 249 amu) were comparable to those of PH46. The presence of 397 amu under negative mode confirmed the existence of the metabolite PH132; while the detection of 381 amu from the positive mode allowed significant insight into the structure of PH132, which suggested that the OH-group was attached onto the aromatic ring within the indene system (Figure 7). The corresponding chromatograms of the MS/MS experiments are shown in Supplementary Figures S10 and S11.
The identity of PH132 generated from the biotransformation screening was confirmed by a series of spiking experiments using LC4 method, including spiking against the in vitro metabolism hepatocyte samples [rat or human hepatocytes (6 μg/ml) spiking with the bio catalytic screening reaction samples being 10 fold more concentrated], and cross spiking between the RLM and Cyps screening samples (same concentration). The EIC at 397 amu was used for clear comparison. The details of these spiking experiments results are provided in Supplementary Figures S12-S17.
Two other tentative metabolites were also observed, PH46-OH’ in one human Cyp-9 (2C9) sample and PH46-diOH in various microbial Cyp samples (Figure 6). In the incubation with Cyp 9 for 24 h, two peaks with the desired mass of 381 amu (PH132-H2O) were observed in the positive mode LC3. The MS/MS experiment revealed that the fragmentation pattern of both peaks was very similar, which suggested one could be the desired metabolite PH132 and one being a different isomer, PH46-OH’. However, due to the low concentration, no further structural conclusion of PH46-OH’was made. The LC-MS and MS/ MS results are given in Supplementary Figures S18 and S19. In some Cyp samples, incubated with microbial P450 enzymes, the peak with 415 amu was observed, being possibly metabolite M3 (PH46-diOH). However, upon further investigation of MS/MS experiments, the fragmentation pattern of the peak with 415 amu had no similarity with that of either PH46 or PH132. It is possible that this observed peak had not originated from PH46, thus not being a metabolite. The relevant chromatograms are given in Supplementary Figures S20 and S21.
Structure elucidation of metabolite PH132 by NMR & HRMS
PH132 was isolated with purity of 95% Rat liver microsomes bio catalytic systems were used in the subsequent scale-up production of PH132 metabolite. Upon reaction work up and purification, The combined samples (from 12 reactions) were dried for 2 d on a high vacuum manifold and 64 mg of PH132 (62% yield) was obtained with purity of 95%, as white powder. PH132 was characterized and its chemical structure was confirmed by NMR experiments using PH46 as reference for comparison (Figure 8). The molecular mass and absolute configurations were also monitored by HRMS and polarimetry. Representative NMR spectra are given below (Figures 9-13). The detailed NMR spectra and chromatograms are provided in Supplementary Figures S22-S28.
1H and 13C NMR data of PH46 were in agreement with the data we previously reported (Figures 9 and 10) [17]. The 1H NMR spectrum of PH132 (Figure 11) shared similar spectral pattern as that of PH46. Structural assignments of PH132 were achieved with combinations of 1D (1H, 13C, DEPTs & NOESY) and 2D (1H-1H COSY, HSQC, HMBC & ROESY) NMR experiments.
The HSQC NMR spectrum of PH46 (blue) overlaying with the HSQC (red) and 1H (green) NMR spectra of PH132 allowed the correlation of the assignments of most proton signals detected in the aromatic region in the 1H spectrum of PH132 (Figure 12). The key feature in this analytical technique was the observation of the signal of the aromatic proton H16 of PH132 at 6.90 ppm being a broad doublet. The 1H-1H COSY spectrum of PH132 confirmed the existence of the aromatic protons H18, H19, H21, H22, H24 and H25 from the correlations observed in Figure 13a.
The position of the double bond on the indene ring was further identified by ROSEY NMR experiments via w-coupling (NOE correlation) between the alkenic proton H19 at 6.90 ppm being a board doublet and the aromatic proton H15 at 7.02 ppm being a multiplet (Figure 13b). As a result, the position of the hydroxyl group of PH132 produced was determined to be at the C17 position. The chirality of PH132 was also measured by optical rotation. A negative value was obtained, which is consistent with PH46. Therefore, PH132 is chemically named as 4-(((1’S,2’S)-1’,6-dihydroxy-1’,3’-dihydro- 1H,2’H-[2,2’-biinden]-2’-yl)methyl)benzoic acid. The details of characterization of PH132 are: 1H NMR (600.1 MHz, DMSO-d6) δ 2.70 (1H, d, J=13.31 Hz, CH2), 32.91 (1H, d, J=15.70 Hz, CH2), 2.94 (1H, d, J=15.70 Hz, CH2), 3.16 (1H, d, J=13.31 Hz, CH2), 3.36 (1H, d, J=22.80 Hz, CH2), 3.47 (1H, d, J=22.80 Hz, CH2), 5.02 (1H, d, J=6.90 Hz, CHOH), 5.75 (1H, d, J=7.66 Hz, OH), 6.27 (1H, s, CH=C), 6.58 (1H, dt, J1=2.74 Hz, J2=8.23 Hz, Ar-H), 6.85-7.36 (8H, 4 x m, Ar-H), 7.69 (2H, d, J=8.28 Hz, Ar-H); 13C NMR (150.9 MHz, DMSO-d6) δ 38.1 (CH2), 38.3 (CH2), 39.2 (CH2), 55.5 (quat. C), 81.0 (CHOH), 111.4 (tert. C), 112.9 (tert. C), 120.4 (tert. C), 124.1 (quat. C), 124.2 (tert. C), 124.4 (tert. C), 126.2 (tert. C), 126.7 (CH=C), 127.5 (tert. C), 128.3 (quat. C), 2 x 128.5 (2 x tert. C), 2 x 129.9 (2 x tert. C), 135.8 (quat. C), 140.4 (quat. C), 144.6 (quat. C), 145.1 (quat. C), 149.9 (quat. C), 154.7 (quat. C), 167.5 (COOH). HRMS (ESI): m/z calculated for C26H21O4 (M-H)+, 397.1440; found 397.1436; [α]D22 = -96.9° (12.8 mg/1 mL, MeOH).
In summary, the in vitro metabolism study of the clinical candidate PH46A were studied by our group. Five metabolites were identified, including M1 (keto-PH46), M2 (PH46-OH or PH132), M3 (PH46-diOH), M4 (keto-glucuronide-PH46) and M5 (glucuronide conjugate- PH46) across species tested; while PH46 was confirmed in all samples at 0 and 240 min incubation time. Overall, all putative human metabolites were present in rat and dog. Metabolism in rat was most similar to human in terms of the metabolites observed. The order of clearance of PH46 was determined to be rat>dog>monkey>human. The species which exhibited the closest clearance values to the human was monkey. The rat produced the highest clearance of PH46 of all the species. Further studies will be carried out to investigate the stereochemistry of each metabolite to determine whether different stereoisomers of the metabolites were present following incubation.
We further developed an efficient biocatalytic method to generate one of the key metabolites, PH132, following an extensive screening of the selectAZyme panel of enzymes. Recombinant human liver Cyps and RLM were identified as being capable of producing desired PH132, which was confirmed by a series of spiking experiments. RLM were selected for use for the subsequent scale-up reaction, and the full chemical structure characterisation of isolated and purified PH132 was confirmed by LC-MS/MS and a complete series of NMR experiments. It was revealed that the position of the OH moiety was at the C17 position locating on the aromatic ring of the indene system. The results obtained from the work are important for understanding the metabolism of PH46A in human and provide important information that may act as a reference for the next stage of pharmacokinetics, in vivo toxicology and clinical pharmacology investigations.
This work was supported by The Wellcome Trust under grant reference no. 067033/Z/02/A, Enterprise Ireland, Charles River (UK) and Almac Science (UK).
The authors declare there is no conflict of interest.