Enzyme Engineering
Open Access
ISSN: 2329-6674
ISSN: 2329-6674
Editorial - (2012) Volume 1, Issue 1
Chlorinated solvents, such as tetrachloroethylene (also referred to Perchloroethylene; PCE) and Trichloroethylene (TCE), are among the most prevalent groundwater pollutants. Its frequent occurrence at contaminated sites is due to its widespread use as an industrial solvent. PCE and its incomplete dechlorination products are known or suspected carcinogens. Therefore, the treatment of PCE bearing wastes and the remediation of PCE contaminated soils and aquifers are a global priority on environmental pollution control.
Although aerobic co-metabolic dechlorination of PCE by tolueneo- xylene monooxygenase of Pseudomonas stutzeri OX1 has been recently reported [1], PCE is recalcitrant under aerobic condition because of its oxidized nature [2].
Hydrogen is generally considered to be a key electron donor to stimulate the reductive dechlorination of chlorinated ethylene [3-5]. Our previous work, Clostridium bifermentans strain DPH-1 has been found to reductively dechlorinate PCE to cDCE (cis-1,2-dichloroethylene) using hydrogen as an electron donor [6]. Generally, the introduction of halorespiring bacteria is expected to be a cost-effective approach to the remediation of PCE-contaminated site [7,8]. Such bacteria can grow by anaerobic respiration, a process that has been referred to as halorespiration or dehalorespiration [6,9-16]. Some pure cultures have been reported to catalyze the reductive dechlorination of PCE to cDCE (Table 1). These organisms belong to species of Dehalospirillum, Desulfomonile, Desulfitobacterium, Dehalobacter and Clostridium [6,9- 16]. Some strains belonging to Dehalococcoides spp. are able to convert PCE to ethylene sequentially (Table 1). He et al. [17] recently identified a Dehalococcoides strain that uses DCE isomers and Vinyl Chloride (VC) but not PCE or TCE as metabolic electron acceptors.
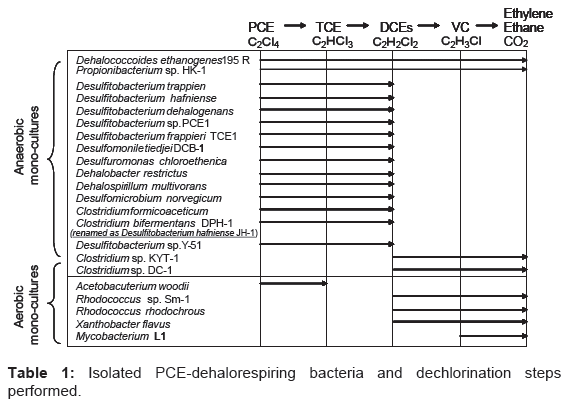 |
Table 1: Isolated PCE-dehalorespiring bacteria and dechlorination steps performed.
PCE dehalogenases have been purified, and their genes cloned, from several bacteria [18-24]. The PCE dehalogenase pceA genes were found to be linked with the pceB genes coding for small hydrophobic proteins containing two or three transmembrane helices [18,25-27], and pceB was assumed to act as a membrane anchor protein to link the dehalogenase to the respiratory chain. The presence of similar PCE genes among different strains strongly indicates that these genes have a mechanism of transfer among these strictly anaerobic bacteria.
Most anaerobic dehalogenases dechlorinate PCE to principally cDCE; however, a novel PCE dehalogenase from Dehalococcoides ethenogens 195 [14] can reductively dechlorinate PCE to ethylene, extensively detoxifying it (Table 1). Aerobic degradation of cDCE by Rhodococcus rhodochrous [28] and Nitrosomonas europaea [29] has been reported. Thus, cDCE accumulation in the anaerobic system can be eliminated by further degradation using such aerobic dehalogenases.
Unlike other dehalogenases from dehalorespiring bacteria, the dehalogenase from strain DPH-1, does not have Fe/S clusters, but exhibits a strong dechlorination activity for PCE as well as several other halogenated compounds. Due to this uniqueness and as a representative PCE-dehalorespiring bacterium, we have reviewed the nature of Clostridium bifermentans DPH-1, through the special focusing on the biochemical organization and genetic regulation of gene encoding PCE dehalogenase followed by the PCE dechlorination.
Two degenerate Primers designed from both ends of the N-terminal amino acid sequence successfully amplified an 81 bp putative region of C. bifermentans PCE dehalogenase. The translated DNA sequence of the probe (81 bp PCR product) matched the predetermined N-terminal protein sequence.
The PCR product was confirmed by DNA sequencing and was used as a probe for gene cloning. Southern hybridization of C. bifermentans genomic DNA cleaved with various restriction enzymes, with probe, with [α-32P] dATP-labelled probe, revealed distinct bands of BglII, ClaI, EcoRI, HindIII. After screening 1720 E. coli DH5α colonies from a genomic sub library (constructed with approximately 4.5-5.5 kb ClaI fragments), we isolated a putative clone (pDEHAL5) containing a 5 kb ClaI insert. Based on the Southern hybridization analysis data (data not shown), it was predicted that a BglII restriction fragment, less than 1.4 kb could harbor the dehalogenase gene. The complete nucleotide sequence of the inserts of the two sub clones, pDEHAF1 and pDEHAF2 (plasmids used) [22], was determined and the gene was identified using the predetermined N-terminal sequence. The gene was found to contain an internal BglII site (Figure 1). From the ATG start codon, a 97 bp of the gene was identified in pDEHAF1 and 1004 bp in pDEHAF2.
The complete nucleotide sequence of pceC was determined. The pceC open reading frame spans from base pair 514 to 1614 (367 amino acids; 39.67 kDa for the mature protein). Hydropathy plot revealed the presence of a discernible signal peptide and methionine was absent from the predetermined amino terminal sequence; indicating post translational processing. The coding region began with a 21 amino acid signal peptide, followed by the predetermined amino terminal sequence (data not shown). Thus, the processed protein (346 amino acids) has a calculated molecular mass of approximately 37.40 kDa, which is identical to that determined by mass-spectroscopic analysis. A putative ribosome binding site (AAAGGA) was found eight bases upstream from the ATG initiation codon. Typical -10 and -35 promoter sequences were found upstream of the coding region. 39 bp downstream of the TGA stop codon is an inverted repeat indicating a rho-independent terminator (Figure 1). The hydropathy profile of the deduced amino acid sequence indicated a major hydrophobic region at the N-terminus, typical of a traditional leader peptide.
PceC is probably a cell-associated extracellular enzyme (peripheral membrane protein) loosely anchored to the cell membrane because it was easily extracted into an aqueous buffer. Moreover, the predetermined N-terminal amino acid sequence started with alanine and the deduced amino acid sequence of pceC contained a discernible signal sequence, indicating a processed peripheral membrane protein. After the four purification steps, the completely purified enzyme was not stable and lost about 50% activity at -30°C. Similar instability was observed with preservation of the enzyme on ice at 4°C. The purified enzyme was therefore not suitable for characterization. This is attributed to the many purification steps and the enzyme’s oxygen sensitivity. Fractions containing the protein impurities, in the semi purified enzyme, showed no PCE dehalogenation activity, indicating the absence of a second dehalogenase in the semi purified enzyme used for characterization.
pceC is a homodimer and differs in molecular size from other reported dehalogenases. PCE dehalogenases of Desulfitobacterium sp. Y-51 [30], Sulfurospirillum multivorans [31] and Dehalococcoides ethenogenes 195 [19] were reported to be monomeric proteins with apparent molecular masses of 57, 60, and 51 kDa, respectively. However, the PCE dehalogenase from strain PCE-S is possibly a homotrimer with an apparent molecular masse of approximately 65 kDa and 200 kDa for monomeric and native forms, respectively.
The most frequently reported dehalogenases consist of a single polypeptide containing one corrinoid cofactor and two iron-sulfur clusters: PCE reductive dehalogenases of S. multivorans [31], Desulfitobacterium sp. strain PCE-S [32], and Desulfitobacterium frappieri TCE-1 [33], ortho-chlorophenol reductive dehalogenases of Desulfitobacterium hafniense [34], Desulfitobacterium dehalogenans [27], and Desulfitobacterium chlororespirans [35] and PCE- and TCEreductive dehalogenases of Dehalococcoides ethenogenes [19]. Two reductive dehalogenases with one corrinoid cofactor but without an iron-sulfur cluster have also been reported: the ortho-chlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1 [36] and the PCE reductive dehalogenase from C. bifermentans DPH-1 [22]. These two proteins are different from all the other dehalogenases already described. The third type of Dehalogenase is a heme protein consisting of two subunits and was isolated only from Desulfomonile tiedjei DCB- 1 [37]. Abiotic dehalogenation of several halogenated compounds was also observed from the heat-inactivated PCE Dehalogenase of S. multivorans and from bacterial transition metal coenzymes vitamin B12 (Co), coenzyme F430 (Ni), and hematin (Fe) [38,39].
The PCE Dehalogenase genes were found to be linked with Open Reading Frames (ORFs) coding for small hydrophobic proteins containing two or three transmembrane helices [18,23,25-27,40,41].
Enzymatic cleavage of halogen-carbon bond (dehalogenation) is a critical step in microbial transformation and mineralization of halogenated aliphatic substances. Dehalogenation, generally, decreases toxicity and, consequently, increases susceptibility of a halogenated molecule to further breakdown. Two strategies have been proposed for the biotreatment of PCE and TCE contamination: (i) complete degradation by reductive dechlorination [14,42], and (ii) by a combination of an aerobic and aerobic systems, in which PCE or TCE is converted to cDCE by anaerobic reductive dechlorination, followed by complete aerobic metabolism of cDCE [43]. The capacity of pceC to effect rapid dechlorination of PCE could be very useful in the proposed two-stage anaerobic and aerobic biotreatment strategy. Mixtures of chlorinated aliphatic substances are often found in polluted environments. However, only a few studies have described the anaerobic transformation of chlorinated hydrocarbons [12,44]. C. bifermentans dehalogenase is unique in that it represents the first characterized anaerobic reductive dehalogenase acting on multiple chlorinated aliphatic molecules. Surprisingly, cDCE was also dechlorinated when added as the initial compound for dechlorination. The product(s) of cDCE dechlorination and the reasons why cDCE as an intermediate product is recalcitrant are not clear. This aspect requires in depth studies to understand the mechanism and product(s).
Genes coding for the first type of reductive dehalogenases have been reported, such as cprA from D. dehalogenans [26,27], pceA from S. multivorans [31] and Desulfitobacterium sp. strain Y51 [24], and tceA from Dehalococcoides ethenogenes [18]. These genes are all closely linked to genes cprB, pcrB, and tceB, respectively, which encode for hydrophobic proteins potentially acting as membrane anchors for the dehalogenases. Villemur et al. [45] isolated genes from D. frappieri PCP-1 that are highly related to cprA and cprB. Furthermore, they also observed several genes coding for putative cprA and pceA in the genomes of D. hafniense DCB-2 and Dehalococcoides ethenogenes. Gene coding for reductive dehalogenases containing corrinoid cofactor but without an iron-sulfur cluster have been reported: crdA, coding for an enzyme mediating the ortho-chlorophenol reductive dehalogenation in D. frappieri PCP-1 [36], and pceC, coding the PCE reductive dehalogenase in C. bifermentans DPH-1 [22]. Both genes and gene products show no similarity with each other and with the first type of reductive dehalogenases.
It is now widely accepted that anaerobic halorespiring bacteria are among the key players in biologic dechlorination processes under anoxic environments. In order to meet these demands recently, a few of PCE Dehalogenase was purified from some halorespiring strains, and cloned like pceA gene. However, details biochemical organization and genetic regulations of this enzyme remain unclear. Literally, function of gene is subjected to physicochemical nature of substrate i.e. arrangement of amino acid sequences. Functionally, chloroethylene dehalogenase is classified into four categories e.g. pceA, vcrA, tceA, and cprA on the basis of substrate specificities. The substrate is recognized by chemical structures or the differences between number and position of the chlorine. The substrate might be able to be expanded by specifying the amino acid at the active center and modifying this might improve reactive efficiency. So, X-ray structural analysis of these enzymes need to be performed. It will be urgent to establish DNA recombination experimental system including the host-vector system of obligatory anaerobe. Next to the numerous technical difficulties of performing genetics on strictly anaerobic bacteria, the tendency of the dehalospiring bacteria to contain multiple reductive dehalogenase gene clusters may attenuate, if not suppress completely, the effect of single gene inactivation. On the other hand, a number of dehalogenase genes are found from the genome of Dehalococcoides ethenogenes 195 [46,47], Desulfitobacterium sp. Y51, and Desulfitobacterium hafniense DCB-2 [48]. Whole genomic analysis of the strain DPH-1 has also been conducting. Thus, the electron transfer system of dehalorespiring bacteria would be clarified in the near future. It is extremely important for considering the origin and the evolution of dehalorespiring bacteria to clarify the function of these genes. The study of dehalorespiring bacteria is able not only to be offered an interesting basic finding but also to be applied to the bioremediation of chlorinated hydrocarbons, and to be greatly expected the progress in the future.