Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2024)Volume 17, Issue 3
Tissues encompass a quality control mechanism that promotes their optimal state. This mechanism, designated cell competition, is characterized by the elimination of suboptimal yet viable cells when they are near healthier cells within the same tissue compartment.
This study explores flower-dependent cell competition and introduces ikebana as a novel player. The differential expression of the flower isoforms labels cells as winners or losers, influencing their fate in diverse contexts, including eye development, traumatic brain injury and Alzheimer's disease. Ikebana, ubiquitously produced in wing imaginal discs and adult brains, modulates loser cell elimination. Reduction of ikebana expression correlates with an increased number of loser cells, while its overexpression in the Alzheimer’s disease model reduces the number of Flower LoseB-positive cells.
We suggest that ikebana protects loser cell elimination, particularly when excessive elimination of loser cells can compromise tissue function. Thus, ikebana might be a potential therapeutic target for modulating flower LoseB expression.
Ikebana; Flower; Cell competition; Drosophila melanogaster
The Darwinian principle of "survival of the fittest" has transcended the animal kingdom to encompass the cellular landscape within their bodies [1]. Current knowledge recognizes that cells within the same tissue compartment engage in a phenomenon known as cell competition, a process elucidated as early as 1975 with the observation of slow-proliferating minute ribosomal mutants being outcompeted by normally proliferating cells [2]. This phenomenon extends beyond mere cell proliferation differences, leading to a broader definition of cell competition; the elimination of viable yet suboptimal cells when juxtaposed with fitter counterparts within the same tissue compartment [3].
Cell competition has been documented across various tissues in organisms ranging from Drosophila and mice to zebrafish and humans, with implications in neurodegenerative diseases and cancer biology [4-10]. The prevailing consensus in the field identifies three major mechanisms of cell competition. First is competition for survival factors, exemplified by decapentaplegic capture during wing imaginal disc development, where cells closer to essential factors or are able to capture them, gain a competitive advantage [11]. Second, mechanical cell competition, where cells compete for space and the resistance to mechanical forces determines their fate, as seen in the elimination of cells converging to the midline in the Drosophila thorax [12-14]. Lastly, cells may compete based on their fitness state, influenced by factors such as ageing or exposure to toxic environments [10,15]. This leads to the elimination of suboptimal cells (losers) and their potential replacement by fitter cells when near healthier (winner) cells. Fitness fingerprints are proteins that recognize and define fitness states and are thought to play significant roles in the execution of cell competition. The fitness fingerprints that have been identified to date include flower, Slit-Robo-Ena, Spatzle-Toll, Stranded at Second and its Receptor Protein Tyrosine Phosphatase 10D (Sas-PTP10D) and flamingo [16-22].
This study delves into flower-dependent cell competition, which relies on the transmembrane protein flower [16]. Although flower has been initially described as a calcium channel, such function appears irrelevant to cell competition [9,23-25]. In Drosophila, flower exists in three isoforms; flower LoseA and flower LoseB, marking loser cells and flower Ubi, marking winner cells [16]. The differential expression of these isoforms determines the fate of a cell as a winner or loser. The expression of flower lose isoforms in Drosophila causes the elimination of suboptimal cells in various contexts, including during eye development, in response to traumatic brain injury and in an Alzheimer’s disease model [10,25-28]. The significance of flower extends beyond Drosophila, with orthologues described in mice and humans [9,29].
In Drosophila, loser cells can experience various fates; if they express Secreted Protein Acidic and Rich in Cysteine (SPARC), they are protected from elimination; on the other hand, if they express the fitness checkpoint azot, they are then marked to die [15,30]. Azot, a predicted calcium-calmodulin, activates hid and the subsequent machinery, leading to apoptosis [15]. However, the mechanisms underlying neighbor recognition as winners or losers, the criteria for selecting loser cells for elimination and the interplay between flower, azot and apoptosis remain areas of ongoing exploration.
This study introduces ikebana as a new participant in the landscape of cell competition, which acts as a key regulator in determining cell fate. In scenarios where competition is intensified, as seen in Alzheimer's disease, increasing ikebana expression reduces the number of cells marked as losers. Our findings position ikebana as a modulator of loser cell elimination, which might be particularly relevant in contexts where excessive labelling of cells as losers could compromise tissue function. With predicted human orthologs, ikebana presents a potential avenue for developing novel therapies to modulate flower expression and cell competition.
Drosophila genetics and experimental setups
Stocks and crosses were kept at 25°C in Vienna standard media with extra yeast. All stocks were obtained from Bloomington Stock Center unless specified. The following Ribonucleic Acid interference (RNAi) lines from Vienna Drosophila Resource Center (VDRC) were used; UAS-ikebana-RNAi (ID 111608, Chr2, viable), UAS-eiger-RNAi (ID 12658, Chr3, viable), UAS-azot-RNAi (ID 7219, Ch3, viable). For the overexpression of ikebana, a UAS-ikebana stock was generated according to standard procedures.
For the super competition assay, the following stocks were used; tub>dmyc>Gal4, UAS-GFP, UAS-white-RNAi, UAS-flower-LoseA/B-RNAi, UAS-lacZ and UAS-diap1. The larvae were given a 17-minute heat shock at 37°C and the vials were placed at 29°C until dissected.
For the wild-type clones in the wild-type background, the following stocks were used; act>y+>Gal4, UAS-GFP and UAS-flowerloseB. The larvae were given an 8-minute heat shock at 37°C and the vials were placed at 29°C until dissected.
For the experiment to understand the effects of ikebana Knockout (KO) in the number of flower LoseB or azot-positive cells, the following stocks were used; white; ikebana{KO}T/ikebana{KO}L; GMR-Gal4, UAS-Aβ42; flower{KO;KI-flowerloseB::mCherry}; azot::mCherry. After hatching, female flies were kept at 29°C until 2 weeks old, when they were dissected.
For the experiment to understand the effects of overexpressing ikebana in the number of Flower LoseB-positive cells, the following stocks were used: GMR-Gal4 and UAS-GFP. After hatching, female flies were kept at 29°C until 2 weeks old, when they were dissected.
For the experiments to understand if Ikebana is a general regulator of apoptosis, the following stocks were used: GMR-Gal4, UAS-eiger and Engrailed-Gal4. The eyes or genitalia were observed in the first week of adulthood.
Ikebana knockout-L generation
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) mediated mutagenesis followed by homologous recombination was performed by the Fly Platform at the Champalimaud Foundation, using the methods highlighted in Baena-Lopez et al. In brief, the upstream guide RNA (gRNA) sequence ACCAACTGCTTGAACCA[GTC] and the downstream gRNA sequence [GCT]ACACCAAGATTTAAGCT were cloned into a pCFD5 vector. The cassette 3xPax3::mCherry contains one attP site, a floxed 3xPax3::mCherry and two homology arms were cloned into pTV3 as a donor template for repair.
CG15098-targeting gRNAs and a donor plasmid were microinjected into embryos nos-Cas9. F1 flies carrying the selection marker 3xPax3::mCherry were further validated by genomic Polymerase Chain Reaction (PCR) and sequenced. CRISPR generates an 1139-bp deletion allele of CG15098, deleting the partial 5’ UTR/3’ UTR and entire Coding Sequence (CDS) of the CG15098 gene and replacing it with cassette 3xPax3::mCherry. This cassette was then removed by Cre-lox recombination, leaving only the attP site and the loxP.
Ikebana knockout-T generation
CRISPR-mediated mutagenesis was performed by WellGenetics Inc. using modified methods of Kondo and Ueda 2013. Briefly, the upstream gRNA sequence GTGAATCCAGAATGCTGTCC[AGG] and the downstream gRNA sequence GGCCAAACGGGAAGCTACAC[TGG] were cloned into U6 promoter plasmid(s) separately. Cassette RMCE-3xPax3::RFP contains two attP sites, a floxed 3xPax3::RFP and two homology arms were cloned into pUC57-Kan as donor templates for repair.
CG15098-targeting gRNAs and hs-Cas9 were supplied in Deoxyribonucleic Acid (DNA) plasmids and a donor plasmid for microinjection into embryos of control strain w [1118]. F1 flies carrying the selection marker 3xPax3::RFP were further validated by genomic PCR and sequencing. CRISPR generates a 1029-bp deletion allele of CG15098, deleting partial 5’ UTR/3’ UTR and entire CDS of CG15098 gene and is replaced by cassette RMCE-3xPax3::RFP. This cassette was then removed by Cre-lox recombination, leaving only the attP sites and the loxP.
Ikebana knockin generation
For the generation of the ikebana{KO; KI-LexA::p65}, the Complementary DNA (cDNA) of LexA::p65 was generated and inserted into a RIVCherry vector and the Knockin (KI) was generated as described in Huang et al., [31]. Primer sequences are available upon request.
Immunohistochemistry and image acquisition
Wing imaginal discs of third-instar larvae were dissected in chilled Phosphate Buffered Saline (PBS), fixed for 20 min in formaldehyde (4% v/v in PBS) and permeabilised with Polybutylene Terephthalate (PBT) 0.4% Triton.
Adult brains were dissected in chilled PBS; the samples were fixated for 30 min in formaldehyde (4% v/v in PBS) and permeabilised with PBT 1% Triton. The wing imaginal discs or brains were then incubated with rabbit Alpha Decapping Protein 1 (α-Dcp1) (1:50) from cell signaling (#9578). Samples were mounted in Vectashield with 4',6-Diamidino-2-Phenylindole (DAPI) (Vectorlabs).
Confocal images were acquired with Zeiss Laser Scanning Microscopy (LSM) 880 using the Plan-Apochromat 20X/0.8 M27 dry objective for the case of the wing imaginal discs and the plan-apochromat 40X/1.4 Oil Differential Interference Contrast (DIC) M27 objective for the case of the adult brains. Maximum intensity projections of the wing imaginal discs or 41-mm-wide images of the adult brains were obtained with Zeiss Zen Black.
To capture images of the adult eyes and genitalia, the Stereomicroscope (Leica S9 I) was used. Flies were either anaesthetized or imaged alive in a CO2 station.
Quantification and statistical analysis
Fiji-ImageJ macros developed in this work, available upon request, quantified the clone areas as the number of flower LoseB, azot or Decapping Protein 1 (Dcp1) positive cells. The areas of the adult eyes were measured in Fiji-ImageJ. For each condition, a minimum of 10 wing imaginal discs, 23 optic lobes and 12 adult eyes were analyzed.
Statistical significance between groups was calculated using the nonparametric Kruskal-Wallis test and Dunn’s test was applied for multiple comparisons between genotypes. Statistical significance between groups was calculated using the Brown-Forsythe and Welch Analysis of Variance (ANOVA) tests and Dunnett’s Test (T3) was applied for multiple comparisons between genotypes.
Ikebana is predicted to interact with flower and is basally produced in the wing-imaginal discs and adult brain
Our understanding of the flower-dependent cell competition pathway remains limited, so our objective was to characterize novel participants in this process. Based on a co-affinity precipitation assay coupled with mass spectrometry, we focused on proteins predicted to interact with Flower, which revealed 34 proteins as potential interactors [32]. From this list, only Mec2 and CG15098 were expected to be transmembrane proteins, which is significant as communication and fitness status labelling likely occur at the plasma membrane. Out of these two proteins, we previously found that only CG15098 is upregulated in loser cells in a microarray analysis designed to identify genes differentially expressed during cell competition [16]. We have named this gene ikebana, drawing inspiration from the Japanese art of flower arrangements. Ikebana has four transmembrane domains, with the -N and -C terminus predicted to be intracellular (Figure 1A) and features two predicted domains; Metal-Transport Protein (Mtp) and Domain of Unknown Function 4728 (DUF4728); placing it within the family of tetraspanin-pasiflora proteins [33]. (A) Predicted configuration of the Ikebana protein at the cell membrane. The green letters a-d indicate the four transmembrane domains represented by the green letters a-d. Model done with Protter; (B) Strategy for the generation of the ikebana{KO}L and, consequently, the ikebana{KO; KI-LexA::p65} transgenic lines. The entire ikebana coding sequence in the genome is removed by CRISPR with two sgRNA. Then the genome is repaired, by homologous recombination, allowing the introduction of the 3xPax3-mCherry selection marker (founder line 1) [31,34]. For the ikebana{KO}L transgenic line, the 3xPax3-mCherry selection marker was removed by Cre-lox recombination. For the generation of the ikebana{KO; KI-LexA::p65} transgenic line, a knockin construct containing the LexA::p65 sequence under the control of the endogenous ikebana promoter, was integrated into the ikebana knockout locus. The pTV3 vector backbone was removed in the final knockin line. (C-D) Expression of Ikebana in the wing imaginal discs of third-instar larvae; (C) and the adult brain; (D) Green Fluorescent Protein (GFP), in green, marks the cells trying to express ikebana and DAPI, in blue, marks the cell nuclei. Scale bars, 50 μm.
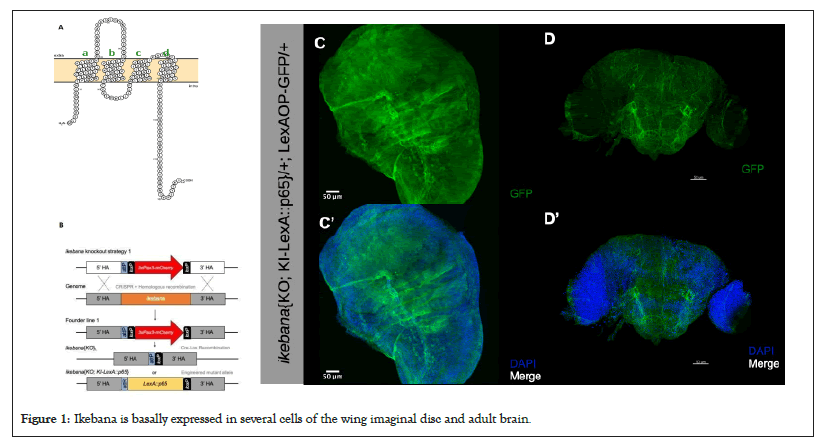
Figure 1: Ikebana is basally expressed in several cells of the wing imaginal disc and adult brain.
To evaluate the expression of ikebana, we employed CRISPR-Cas9 technology, coupled with homologous recombination to generate a genetic ikebana Knockout (KO) line and introduce a genetic marker to allow its identification [31,34]. In this ikebana KO line, we inserted a LexA::p65 sequence under the control of the ikebana endogenous promoter, which, in conjunction with LexAop-GFP, facilitated the specific labelling of cells trying to express ikebana (Figure 1B). We found that ikebana exhibits endogenous widespread expression across third-instar larvae wing imaginal discs and within the adult brain (Figure 1C).
Ikebana partially protects loser cells from elimination
To elucidate the role of Ikebana in cell competition, we conducted experiments to investigate the consequences of modulating the expression of ikebana in loser clones. For this purpose, we used the super competition assay, which relies on the tub>dmyc>Gal4 cassette [35]. Upon activating a heat shock Flippase, this system facilitates the recombination of Flippase Recognition Target (FRT) sites, allowing the expression of both GFP and a construct of interest. By manipulating the heat shock duration, we generated GFP-marked clones scattered within cells carrying an additional copy of dmyc. This supplementary myc copy designates the background cells as winners, while the clones lacking this additional copy are losers whose elimination can be prevented by downregulating the flower lose isoforms or overexpressing Death-Associated Inhibitor of Apoptosis 1 (diap1).
Contrary to downregulating flower LoseA/B, downregulating ikebana in the loser clones did not block their elimination at 72 h After Clone Induction (ACI), as shown in Figures 2A-2D. However, the overexpression of ikebana in the loser clones significantly reduced their elimination compared to the negative control at 72 h ACI (Figures 2E-2H). Notably, this protective effect was not as pronounced as when overexpressing diap1, suggesting that Ikebana confers partial protection to loser cells from elimination (Figure 2). (A-C) Super competition assay in the wing imaginal discs of third instar larvae in which loser wild-type clones (GFP) are outcompeted by dmyc overexpressing cells. UAS-white-RNAi (A), UAS-flower lose A/B-RNAi; (B) and UAS-ikebana-RNAi; (C) Are expressed in the loser clones 72 h after clone induction (ACI). DAPI is represented in blue. Scale bars, 100 mm; (D) Quantification of the clone area over the total area of the wing pouch when UAS-white-RNAi, UAS-flower LoseA/B-RNAi and UAS-ikebana-RNAi are expressed in the loser clones 72 h ACI. We normalized the ratio at 12 h ACI (data not shown) and subsequently, the 72 h time point is normalised relative to their respective conditions at 12 h; (E-G) Super competition assay in the wing imaginal discs of third instar larvae in which loser wild-type clones (GFP) are outcompeted by dmyc overexpressing cells. UAS-lacZ (A) UAS-diap1; (B) and UAS-ikebana; (C) are expressed in the loser clones 72 h ACI. DAPI is represented in blue. Scale bars, 100 mm; (H) Quantification of the clone area over the total area of the wing pouch when UAS-lacZ, UAS-diap1 and UAS-ikebana are expressed in the loser clones 72 h ACI. We normalised the ratio at 24 h ACI (data not shown) and subsequently, the 72 h time point is normalized relative to their respective conditions at 24 h.
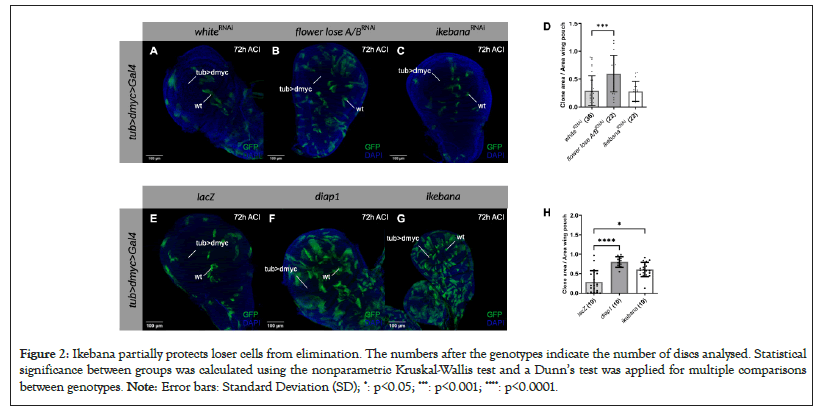
Figure 2: Ikebana partially protects loser cells from elimination. The numbers after the genotypes indicate the number of discs analysed. Statistical significance between groups was calculated using the nonparametric Kruskal-Wallis test and a Dunn’s test was applied for multiple comparisons between genotypes. Note: Error bars: Standard Deviation (SD); *: p<0.05; ***: p<0.001; ****: p<0.0001.
To ascertain that this effect is specific to cell competition, we conducted experiments to exclude the possibility that ikebana is a general apoptosis regulator. First, we expressed eiger-cell death initiator via Jun N-terminal Kinase (JNK) signaling in the eye, which resulted in a reduced eye phenotype [36]. We then examined whether ikebana could rescue the normal phenotype. Additionally, considering that the rotation of the fly terminalia during development is apoptosis-dependent [37], we investigated whether overexpressing or downregulating ikebana in this region would impact normal terminalia rotation, manifesting as incomplete terminalia rotation in the adult fly. Results show that modulating ikebana expression does not interfere with the eye phenotype or terminalia rotation, indicating that Ikebana is not a general regulator of apoptosis and that the effects observed in clones are indeed attributable to cell competition.
Ikebana regulates flower LoseB expression
Given the basal expression of ikebana in the wing imaginal discs of the third instar larvae, we explored whether downregulating its expression with ikebana RNAi can induce cell competition. Using the act>y+>Gal4 cassette, we generated GFP-labeled wild-type clones surrounded by cells expressing an additional copy of yellow, which are also wild type in the context of cell competition. As a negative control, we expressed white-RNAi in the clones, which is not predicted to interfere with cell competition and as a positive control; we overexpressed flower LoseB, which will confer a loser state to the clones, resulting in their elimination over time. Competition assay in the wing imaginal discs of third instar larvae in which wild-type clones (GFP) are in a background of yellow expressing cells (Figure 3) [16]. (A) UAS-white-RNAi; (B) UAS-flowerloseB; (C) UAS-ikebana-RNAi are expressed in the wild type clones 72 h ACI. DAPI is represented in blue. Scale bars, 100 mm; (D) Quantification of the clone area over the total area of the wing pouch UAS-white-RNAi, UAS-flower-LoseB and UAS-ikebana-RNAi are expressed in the loser clones 72 h ACI. We normalised the ratio at 24 h ACI (data not shown) and subsequently, the 72 h time point is normalised relative to their respective conditions at 24 h; (E) Strategy for the generation of the ikebana{KO}T transgenic line. The entire ikebana coding sequence in the genome is removed by CRISPR with two sgRNA [34]. Then the genome is repaired, by homologous recombination, allowing the introduction of the 3xPax3-RFP selection marker (founder line 2) [31]. The 3xPax3-RFP marker was removed by Cre-lox recombination; (F-I) Expression of Flower LoseB and Dcp1 in ikebana KO adult optic lobes; (F) Optic lobe of control flies; ikebana{KO}; GMR>Aβ42; (I) GMR>Aβ42, ikebana{KO}. Flower LoseB is marked in red; α-Dcp1 is marked in magenta; DAPI, in blue, marks the cell nuclei. 41 mm image projections. Scale bars, 50 mm; (J) Quantification of the number of Flower LoseB positive cells in the optic lobes normalised against the control (ctr)+; (K) Quantification of the number of Dcp1 positive cells in the optic lobes normalised against the ctr+; (L-O) Expression of Flower LoseB in the adult optic lobes when ikebana is overexpressed; (L) Optic lobe of control flies overexpressing GFP; (M) ikebana; (N) flies expressing the Aβ42 peptide in the GMR region, also overexpressing GFP; (O) ikebana. Flower LoseB is marked in red; DAPI, in blue, marks the cell nuclei. 41 mm image projections. Scale bars, 50 mm; (P) Quantification of the number of Flower LoseB positive cells normalised against the control GFP in the ctr scenario.
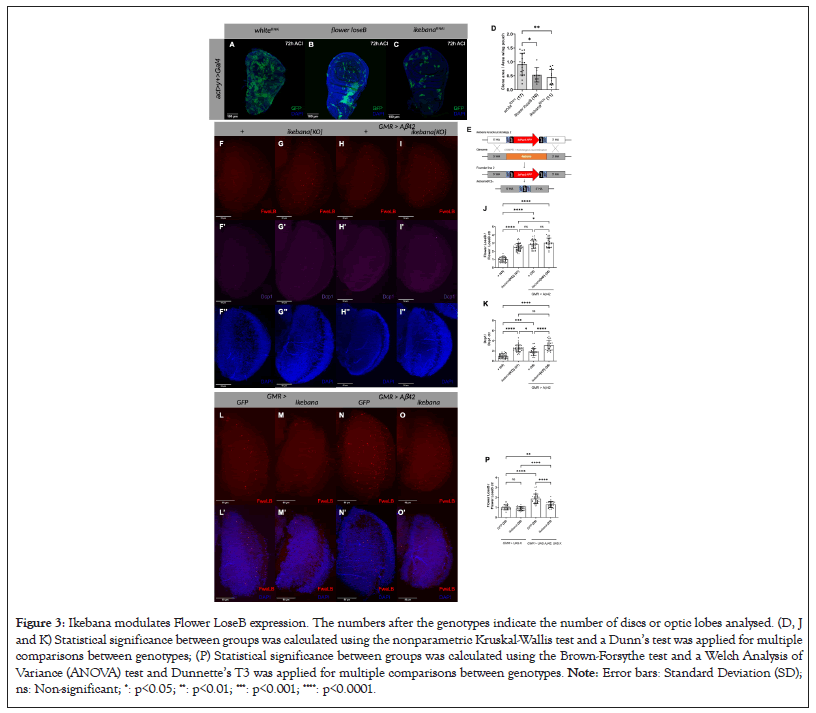
Figure 3: Ikebana modulates Flower LoseB expression. The numbers after the genotypes indicate the number of discs or optic lobes analysed. (D, J and K) Statistical significance between groups was calculated using the nonparametric Kruskal-Wallis test and a Dunn’s test was applied for multiple comparisons between genotypes; (P) Statistical significance between groups was calculated using the Brown-Forsythe test and a Welch Analysis of Variance (ANOVA) test and Dunnette’s T3 was applied for multiple comparisons between genotypes. Note: Error bars: Standard Deviation (SD); ns: Non-significant; *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001.
At 72 h ACI, ikebana-RNAi expression resulted in a reduced clonal area compared to the white-RNAi control. Such reduction of the clonal area was even more pronounced than the one observed with the overexpression of flower LoseB (Figures 3A-3D). This suggests that decreasing ikebana expression is sufficient to induce a loser state, although more experiments are required to prove that this is due to cell competition.
We then asked whether Ikebana regulates the loser state of a cell by influencing Flower expression. Using a FlowerLoseB::mCherry reporter line, we assessed its production in an ikebana KO scenario (Figures 3E-3K). For the ikebana KO condition, we crossed the ikebana Knockout Target (KOT) with the ikebana Knockout Line (KOL), a transheterozygous allelic version, because we observed the presence of off-target effects in the ikebana KOL line (data not shown). In the optic lobe of ikebana KO flies, we found a 2.5-fold increase in Flower LoseB-positive cells compared to the control wild type for ikebana (Figure 3J). This rise in the number of loser cells was accompanied by increased Dcp1-positive cells (Figure 3K), meaning these cells were actively being eliminated.
We then tested an Alzheimer’s disease model scenario where we expected more loser cells [10]. Intriguingly, in this scenario, removing ikebana did not further increase the number of loser cells in the optic lobe (Figure 3J). However, it did increase the number of dying cells (Figure 3K), as measured via Drosophila Caspase 1 (Dcp-1) positivity. We also examined whether the same happened for azot expression and indeed, the absence of ikebana is sufficient to increase the number of azot-positive cells in basal cell competition but not in an Alzheimer’s disease model. These data suggest that the absence of ikebana alone is often sufficient to increase the number of loser cells. However, in an Alzheimer’s disease model, ikebana prevents cell elimination without affecting flower LoseB expression, as seen by the increase in dying cells in the ikebana KO scenario without an increase in the number of loser cells.
Lastly, we investigated whether overexpressing ikebana in the optic lobe, using the Glass Multiple Reporter (GMR) driver, would decrease the number of flower LoseB-positive cells (Figures 3L-3P). Under basal levels of cell competition, overexpressing ikebana did not affect the number of flower LoseB-positive cells. However, in an Alzheimer’s disease model, overexpressing ikebana in the GMR region led to a 0.7-fold decrease in the number of cells producing flower LoseB::mCherry compared to the GFP control (Figure 3P). This suggests that high levels of ikebana can reverse the low fitness status of cells, indicating that ikebana is sufficient to partially block the loser fate specification in the Alzheimer’s disease model.
Ikebana, anticipated as a transmembrane protein, which physically interacts with flower, emerges as a pivotal contributor to cell competition dynamics. Using a clonal assay, we observed that localized reduction of ikebana expression within clones results in a diminished clonal area, similar to local overexpression of flower loseB, which forces the cells into a loser state. Given the seemingly ubiquitous expression of ikebana in wing imaginal discs, we propose that, for a cell to adopt a loser state, it must first downregulate ikebana. We hypothesize that this decrease in ikebana production will promote the production of flower LoseB, leading to cell elimination. In adult brains, the absence of ikebana proves sufficient to increase the number of cells positive for flower LoseB or azot, further supporting this hypothesis. In scenarios anticipating a high number of loser cells, as in the Alzheimer’s disease model, overexpressing ikebana correlates with a decrease in the number of flower LoseB-positive cells, implying that ikebana can revert the low fitness of the cells; a refined mechanism which likely operates to selectively eliminate the necessary number of losers, preserving tissue function. In such cases, ikebana can be re-expressed in loser cells, reverting their loser state by decreasing flower LoseB expression and sparing selected loser cells from elimination (Figure 4).
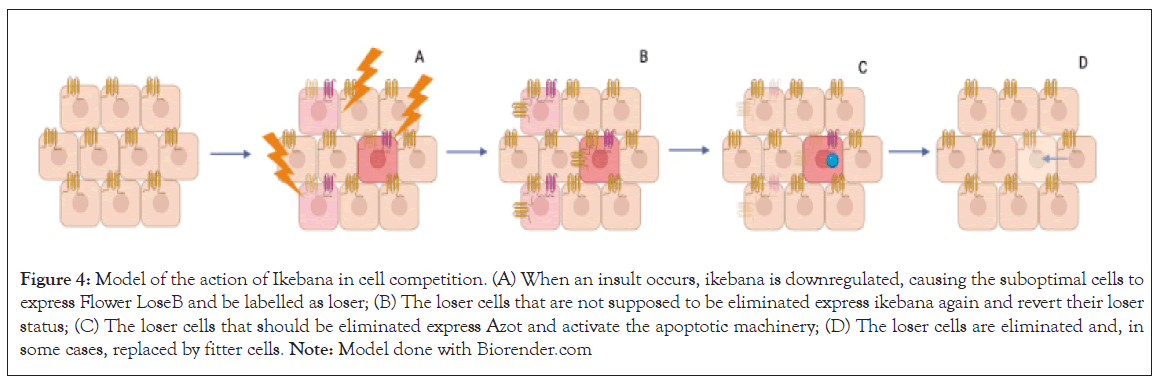
Figure 4: Model of the action of Ikebana in cell competition. (A) When an insult occurs, ikebana is downregulated, causing the suboptimal cells to express Flower LoseB and be labelled as loser; (B) The loser cells that are not supposed to be eliminated express ikebana again and revert their loser status; (C) The loser cells that should be eliminated express Azot and activate the apoptotic machinery; (D) The loser cells are eliminated and, in some cases, replaced by fitter cells. Note: Model done with Biorender.com.
To strengthen the model, additional experiments are needed to exclude interference with cell proliferation. Overexpressing or downregulating ikebana in specific regions and comparing sizes against a control without ikebana manipulation, coupled with immunostaining for cell death in clones, would bolster the robustness of our conclusions. Although these experiments were not conducted, observational evidence using the actin Galactose-Responsive Transcription Factor GAL4 (Gal4) driver indicates normal development time and adult fly size when manipulating ikebana (data not shown). This supports our conviction that the observed effects are due to cell competition rather than mere differences in proliferation rates.
Ikebana joins a growing list of proteins, such as SPARC, implicated in protecting loser cells during Flower-dependent cell competition [30]. However, SPARC does not belong to the Flower-dependent cell competition canonical pathway because manipulating flower in loser or wild-type clones does not change the levels of SPARC [30]. On the contrary, Ikebana is proposed to interact directly with Flower. Additionally, SPARC is exclusively expressed in loser cells to prevent their elimination, whereas ikebana appears ubiquitously expressed.
Interestingly, Ikebana was previously identified as a member of the tetraspanins-pasiflora family of proteins, characterised by four transmembrane domains and conserved regions [33]. In Drosophila, only two other proteins, pasiflora and fire exit, are characterised, albeit in embryos and their role in cell competition remains unexplored [33,38]. This protein family extends to humans, including the lysosomal-associated proteins LAPTM4A and LAPTM4B. Ikebana shares more structural similarities with Lysosomal-Associated Protein Transmembrane 4 Alpha (LAPTM4A) [33]. This human protein, associated with the clearance of proteins from the plasma membrane via endocytosis and implicated in inducing multidrug resistance in Saccharomyces cerevisiae, presents a potential avenue for therapeutic exploration [39-41]. Future work is needed to confirm LAPTM4A as the human orthologue of ikebana and test if it is involved in the clearance of Flower lose isoforms from the cell membrane. If so, manipulation of LAPTM4A could open therapeutic avenues to prevent the unnecessary elimination of loser cells.
Our research proposes ikebana as a novel transmembrane protein involved in cell competition. As a predicted flower-interacting protein, ikebana shares structural similarities with tetraspanins-pasiflora proteins and is basally expressed in various tissues. While not a general apoptosis regulator, ikebana plays a significant role in protecting loser cells from elimination, potentially by modulating flower LoseB signaling. This function suggests that Ikebana fine-tunes the loser state during active cell competition, ensuring optimal elimination of loser cells without compromising tissue integrity. The identification of LAPTM4A as a potential human orthologue of ikebana opens avenues for further research in mammalian systems, although experimental validation is necessary. These findings contribute significantly to our understanding of cell competition mechanisms and may have implications for developmental biology and cancer research. Future studies should focus on elucidating the precise molecular interactions between ikebana and flower, as well as exploring its potential role in human cellular processes.
M.M.R., A.G.G. and E.M. designed the experiments. M.M.R. performed and analyzed the experiments. A.G.G. helped with image acquisition and statistical analysis. C.B.P. helped with data analysis and design of the transgenic constructs. C.F.C., I.A. and M.S.E obtained preliminary data. B.H. developed the UAS-ikebana transgenic flies. M.M.R. wrote the manuscript.
We thank Bloomington Stock Center for flies; WellGenetics Inc. for the generation of the ikebana KOT; the technicians at the Champalimaud Fly Platform for support with stock maintenance; the MTT platform for support in the generation of transgenic flies, the ABBE platform for microscopy support and Andrea Spinazzola for his suggestions and comments on the manuscript. M.R was supported by an FCT-Fundação para a Ciência e a Tecnologia-PhD studentship (SFRH/BD/138537/2018). This study was supported by Portuguese national funds, through FCT in the context of the project UIDB/04443/2020 and the European Research Council (Consolidator Grant to E.M.: ‘‘Active Mechanisms of Cell Selection: From Cell Competition to Cell Fitness’’). Fly and MTT platforms were funded by the research infrastructure CONGENTO, co-financed by Lisboa Regional Operational Programme (Lisboa2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) and Fundação para a Ciência e Tecnologia (Portugal) under the project LISBOA-01-0145-FEDER-022170. The Portuguese Platform of Bioimaging funded ABBE platform-LISBOA-01-0145-FEDER-022122.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Marques-Reis M, Gutierez-Garcia A, Clara CF, Argudo I, Eggel MS, Hauert B, et al. (2024). Characterizing the Modulatory Role of Ikebana in Flower-Dependent Cell Competition. J Proteomics Bioinform. 17:671.
Received: 02-Aug-2024, Manuscript No. JPB-24-33331; Editor assigned: 05-Aug-2024, Pre QC No. JPB-24-33331 (PQ); Reviewed: 19-Aug-2024, QC No. JPB-24-33331; Revised: 26-Aug-2024, Manuscript No. JPB-24-33331 (R); Published: 02-Sep-2024 , DOI: 10.35248/0974-276X.24.17.671
Copyright: © 2024 Marques-Reis M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.