Research Article - (2015) Volume 1, Issue 1
Characterization and Evaluation of Polycyclic Aromatic Hydrocarbon (PAH) Degrading Bacteria Isolated from Oil Contaminated Soil
*Corresponding Author: Sibi G, Department of Biotechnology, Indian Academy Degree College, Centre for Research and Post Graduate Studies, Bengaluru - 560 043, Karnataka, India, Tel: +91 99864 52875 Email:
Abstract
Polycyclic aromatic hydrocarbons (PAHs) are common environmental pollutants and biodegradation using microorganisms is the preferred and major route of PAH removal from contaminated environments. This study investigated the bacterial degradation of petrol and diesel in liquid media that were isolated from oil contaminated soils by enrichment technique. The isolates could use petrol and diesel as their sole carbon and energy source in Bushnell Hass Mineral Salts (BHMS) medium at 2% (v/v) concentration. A total of eight isolates were selected and characterized by using a variety of phenotypic and morphologic properties. Two isolates each showed highest growth in petrol and diesel containing media during screening were selected and characterized using 16S RNA sequencing. Molecular identification of the isolates assigned them to Achromobacter sp. and Pseudomonas aeruginosa. The selected isolates degraded petrol and diesel up to 31.9% and 34.4% respectively. This study indicates that the contaminated soil samples contain a diverse population of PAH-degrading bacteria and the use of Achromobacter sp. and Pseduomonas aeruginosa has the potential for bioremediation of PAH contaminated sites.
Keywords: Biodegradation; Polycyclic aromatic hydrocarbons; Achromobacter; Pseudomonas; Petrol, Diesel degradation
Introduction
Hydrocarbons play a special role amongst the contaminants polluting the environment due to their wide-scale distribution and hazardous physicochemical and biological properties. Hydrocarbons enter into the environment through waste disposal, accidental spills, losses during transport and storage. Polycyclic aromatic hydrocarbon (PAHs) compounds are among the most toxic components to plants and animals [1,2]. Petroleum hydrocarbons pose as a globally environmental pollutant [3] because of their hydrophobic nature and low volatility [4]. Petroleum constituents such as diesel oil are carcinogenic, mutagenic and potent immunotoxicants [5,6] Despite volatilization, leaching, chemical and photo oxidation are often effective in reducing the environmental level of PAHs [7], bioremediation using biological processes to ameliorate hydrocarbons from environment involves no secondary contamination and offers an effective technology for the treatment of oil pollution [8]. Besides, physicochemical treatments to the remediation of hydrocarbons are expensive and laborious [9].
Bioremediation of soil contaminated with petroleum oil has been commonly described globally [10-13] and is usually the preferred and major route of PAH removal from contaminated environments. Hydrocarbon degradative process is widely distributed among numerous genera or taxa [14] and biodegradation of hydrocarbons using microorganisms has been established as an efficient, economic and versatile approach [15-19]. Microorganisms degrade hydrocarbon contaminants and utilize the resulting compounds as nutrients and energy sources for growth and reproduction [20] and are depending on the chemical nature of the compounds and on environmental determinants [21]. The low aqueous solubility of PAHs makes them poorly available for microbial utilization [22] and identification of novel bacteria for the biodegradation of PAH in oil contaminated area is the need of the hour. The main objective of the present study was to characterize PAH degrading bacteria inhabiting the oil contaminated sites. From the samples, bacterial strains able to degrade petrol and diesel were isolated and characterized. Among the isolates, two efficient strains were selected and their degradation capacities of PAH compounds were evaluated.
Materials and Methods
Study cites
Samples of soil for this study were collected from two cites; oil station in Bangalore (13.04°N, 77.64°E) and Mumbai (19.39°N, 72.84°E).
Sample collection
Subsurface soils contaminated with petrol and diesel oil were picked from study sites in pre-sterilized sample bottles, labelled and transported. The samples were stored at -20°C in the Biotechnology laboratory at the Indian Academy Degree College, Bangalore.
Isolation and enrichment of PAH degrading bacteria
For culture enrichment, 1 g soil was inoculated into 100 ml of Bushnell Hass Mineral Salts (BHMS) medium (MgSO4 .7H2O - 0.2 g, CaCl2- 0.02 g, KH2PO4- 1.0 g, K2HPO4 - 1.0 g, NH4NO3 - 1.0 g, 2 drops of 60% FeCl3, pH - 7.0) supplemented with 0.5% petrol and diesel (v/v) as the sole carbon source. The mixture was incubated at 37°C at 130 rpm for 5 days. Bacteria from this culture were enumerated by total plate count method using serial dilution technique on BHMS agar medium supplemented with petrol and diesel. The plates were incubated aerobically at 37°C for 48 h. The criterion for selection of petrol and diesel degrading strains was enhanced growth on plates with added test compound compared to the control plates without added substrate.
Phenotypic identification of the bacterial isolates
The bacterial colonies that grew on BHMS agar plates were subcultured on nutrient agar for pure culture preparation and identification. Eight bacterial isolates were identified on the basis of their colonial and cellular morphology; and biochemical characteristics. The colony colour, margin, form and elevation of the isolates were noted. Gram staining and biochemical tests including; indole, methyl red, Voges-Proskauer, catalase, oxidase, urease test, glucose fermentation and growth on King’s media were also performed to identify the pure colonies. The pure colonies were preserved in glycerol broth (25% v/v) and for the day to day experiments, the bacteria were maintained on nutrient agar plates at 4°C in a refrigerator and sub-cultured at an interval of two weeks.
Screening of efficient PAH degrading bacteria
The efficacy of polycyclic aromatic hydrocarbon degradation was studied by gravimetric assay [23,24]. The bacterial isolates were inoculated into 100 ml of BHMS broth was added with 2% of the respective carbon sources (petrol and diesel) and the flasks were incubated for 5 days at 37°C at 125 rpm. After appropriate incubation, bacterial activities were stopped by adding 1N HCl. For extraction of oil, 50 ml of culture broth was mixed with 50 ml of petroleum ether:acetone (1:1) in a separating funnel and was shaken vigorously to get a single emulsified layer. Acetone was then added to it and shaken gently to break the emulsification, which resulted in three layers. The top layer containing petroleum ether mixed with oil and acetone was passed through anhydrous sodium sulphate followed by evaporation on a water bath. The gravimetric estimation of residual oil left after biodegradation was made by weighing the quantity of oil in a tarred beaker and the percentage of petrol and diesel oil degraded was determined as per the standard method [25]. All these screening experiments have done in triplicate. The average value of triplicate and standard error was calculated by Microsoft XL 2007. The isolates which showed better PAH degradation during the incubation were selected for a detailed analysis.
Genomic amplification of 16S rRNA
Genomic DNA was extracted from overnight grown bacterial cells using InstaGeneTM Matrix Genomic DNA isolation kit. The 16S ribosomal (rRNA) gene from the genomic DNA was amplified by PCR using the following primers; 5’- AGAGTTTGATCMTGGCTCAG-3’ and 5’- TACGGYTACCTTGTTACGACTT-3’ corresponding to the forward and reverse primers respectively. The amplification was done by initial denaturation of 94°C for 2 min followed by 35 amplification cycles of 94°C for 45 s, 55°C for 60 s; 72°C for 60 s and final extension at 72°C for 10 min in MJ Research Peltier Thermal Cycler. The purified PCR product was purified using Montage PCR Clean up kit (Millipore) and the product was sequenced using the 518F/800R primers.
DNA sequencing
Sequencing reactions were performed using a ABI PRISM® BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq® DNA polymerase (FS enzyme) (Applied Biosystems). Single-pass sequencing was performed on each template using 5’- GGATTAGATACCCTGGTA-3’ and 5’- CCGTCAATTCMTTTRAGTTT-3’ primers. The fluorescent-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems).
Analysis of sequence data
The 16S rRNA gene sequence was compared with those from Genbank using n-BLAST program. A phylogenetic tree was constructed by the neighbour-joining method using MUSCLE 3.7 and PhyML 3.0 aLRT program [26]. For tree rendering, Tree Dyn 198.3 was used [27].
Results
Isolation of PAH degrading bacteria
Enrichment cultures initiated in BHMS medium containing 1% (v⁄v) petrol and diesel crude oil as the carbon and energy became turbid and the oil layer became clear indicating the degradation of the compounds. Colonies that have shown enhanced growth on BHMS agar plates added with petrol and diesel compared to control plates were selected. Five colonies from petrol degradation (P1, P2, P3, P4, P5) and three colonies from diesel degradation (D1, D2, D3) were picked and used for further characterization.
Phenotypic identification of PAH degrading bacterial isolates
The colony characteristics of the isolates are summarized in Table 1. The colonies were circular, smooth, elevated and 0.4-1.0 mm in diameter. All the strains were able to grow at 37°C and no pigmentation was observed. The cell morphology and biochemical characteristics are represented in Table 2.
| Isolate | Size | Shape | Colour | Margin | Elevation | Pigmentation | Opacity |
|---|---|---|---|---|---|---|---|
| P1 | 0.5 mm | Circular | Green | Undulate | Convex | Negative | Translucent |
| P2 | 0.8 mm | Circular | Green | Undulate | Convex | Negative | Translucent |
| P3 | 0.7 mm | Circular | White | Lobate | Convex | Negative | Translucent |
| P4 | 0.4 mm | Circular | Green | Lobate | Convex | Negative | Translucent |
| P5 | 0.6 mm | Circular | White | Lobate | Convex | Negative | Translucent |
| D3 | 1.0 mm | Circular | White | Undulate | Convex | Negative | Translucent |
| D4 | 0.8 mm | Circular | White | Undulate | Convex | Negative | Translucent |
| D5 | 0.9 mm | Circular | White | Undulate | Convex | Negative | Translucent |
Table 1: Phenotypic characters of the bacterial isolates.
| Isolate | Gram reaction and cell shape | Catalse test | Oxidase test | Indole test | MR test | VP test | Glucose fermentation | Urease test | Growth on | |
|---|---|---|---|---|---|---|---|---|---|---|
| King’s B | King’s A | |||||||||
| P1 | - rods | + | + | - | - | - | - | - | + | + |
| P2 | - rods | + | + | - | - | - | - | - | + | + |
| P3 | - coccobacilli | + | + | - | - | - | - | - | - | + |
| P4 | - rods | + | + | - | - | - | - | - | + | + |
| P5 | - coccobacilli | + | + | - | - | - | - | - | - | + |
| D3 | - coccobacilli | + | + | - | - | - | - | - | + | + |
| D4 | - coccobacilli | + | + | - | - | - | - | - | + | + |
| D5 | - rods | + | + | - | - | - | - | - | - | + |
Table 2: Gram’s staining and biochemical parameters of the bacterial isolates.
PAH degrading ability of the isolates
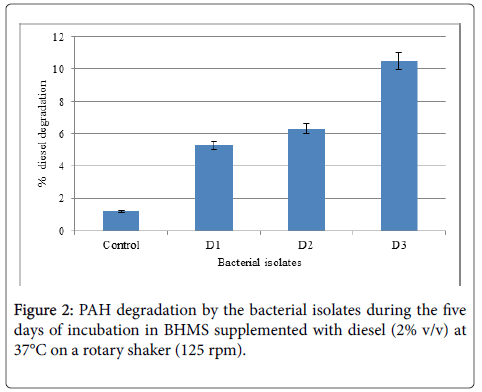
Preliminary biodegradation assay was carried out to determine the PAH degradation capabilities of the indigenous bacterial isolates from oil contaminated environments. Petrol and diesel were added separately at 2% (v/v) to BHMS medium and the incubation was carried out for a period of 5 days for PAH degradation (Figure 1 and 2).
Isolates that exhibited highest degradation of PAH and were selected as efficient strains (P3 and D3) and the degrading ability was analyzed gravimetrically at 5, 10, 15 days of incubation period. The strain P3 was able to degrade petrol with increasing incubation period by recording maximum degradation (31.97%) at the end of 15 days incubation (Figure 3). The degradation rate was minimal till 10 days growth and the degradation rate was 88% increased at the end of incubation period. On the other hand, D3 strain exhibited higher degradation rate at 10th day growth period (34.4%). However, the degradation was significantly reduced with 8.16% degradation at the end of 15 days incubation which was lesser than 10 days degradation period (Figure 4).
Genotypic identification
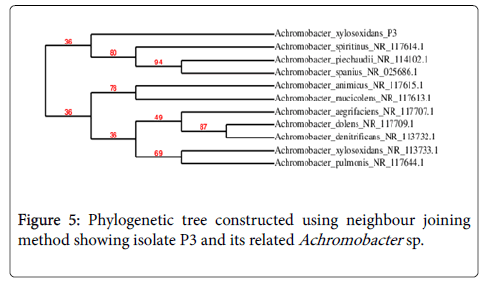
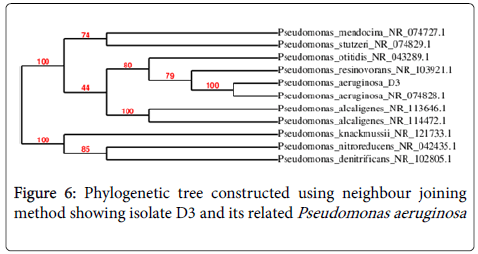
The 16S rRNA gene sequences of P3 and D3 were accessed from the public GenBank data bases using the n-BLAST program. P3 which was Gram negative cocco bacilli was 94% identical to Achromobacter , suggesting that P3 was a strain of this species (Fig-5). D3 was Gram negative coccobacilli and 100% identical to Pseudomonas aeruginosa (Figure 6).
Discussion
Polycyclic aromatic hydrocarbons (PAHs) contain two or more fused aromatic rings in linear, angular, or cluster arrangements [28,29] and are widespread organopollutants present in crude oil and fossilfuel combustion products. Various new regulations have been introduced to control the environmental risks caused by petroleum products and research on remediation of contaminated soils is being increased [30]. The use of microorganisms to eliminate hydrocarbons from contaminated sites has a great potential [31-33]. Certain microorganisms use oil pollutants and microbial degradation of PAH has become the main mechanism for hydrocarbon derived pollution in the environment. Petroleum hydrocarbons are degraded not only by bacteria but by fungi, yeast and microalgae as well [34]. However, bacteria play an essential role in hydrocarbon degradation. In the present work, PAH degrading bacteria were isolated from soil in two different sites using enrichment culture procedure. A total of eight bacteria were isolated and preliminary screened for PAH degradation on BHMS media with 2% hydrocarbon as the sole carbon source individually (petrol and diesel). PAH degraders should use PAHs as sole carbon and energy source for use in bioremediation. This is important for minimizing the production of toxic, water-soluble degradation by-products and reducing the risk of isolates failing to survive at contaminated sites due to the lack of suitable growth substrates. Both the isolates from this research were able to grow well in the presence of petrol and diesel and exhibited significant degradation.
Novel molecular techniques have been extremely valuable in exploring the diversity of microbiota despite culture-based microbiological methods have provided important information about the microbial diversity [35]. The isolates were phenotypically identified and bacterial strains which exhibited relatively higher degradation were selected as most active strains. The tentative taxa and phylogenetic affiliation of the 16S rRNA of purified bacterial isolates were amplified by PCR and the bacterial 16S rRNA sequences were aligned with Blast search of NCBI databases. Partial 16S rRNA gene sequencing and database homology search for the isolates revealed their tentative close relationship to members of Achromobacter sp. and Pseudomonas aeruginosa.
Petroleum hydrocarbons can be degraded by microorganisms aerobically [36-38] and are readily isolated from oil contaminated sites [39]. Using natural populations of microorganisms for the removal of petroleum and other hydrocarbon pollutants from the environment is cheaper than other remediation technologies [40]. A wide range of hydrocarbon concentrations (0.5 - 6%) were used in biodegradation studies and varying growth was obtained [41-45]. In this study 2% (v/v) was used as carbon and energy source for the degradation. In this study, based on preliminary screening of biodegradation, two native bacterial species namely Achromobacter and Pseudomonas aeruginosa were isolated. The experiments were carried out for a period of 15 days and the degrading ability of the isolates were gravimetrically determined at every 5 days interval. In general, the degradation rate was increased with incubation time and the results demonstrated that Achromobacter sp. have the greatest ability to degrade petrol while Pseudomonas aeruginosa demonstrated the greatest degrading ability on diesel. The rate of petrol degradation was maximum (31.9%) at the end of 15 days interval period by Achromobacter . Recently, Achromobacter sp has been reported to utilize polycyclic aromatic hydrocarbons [46-50]. PAH degradation by Pseudomonas strains is well characterized [51]. P. aeruginosa is a good candidate for bioaugmentation of petroleum contaminated soils [52] and its degradation of n-alkanes in diesel oil is reported [53]. Diesel oil is a complex mixture of alkanes and aromatic compounds that frequently are reported as soil contaminants [54]. Diesel degrading strain of Pseudomonas aeruginosa capable of growing in the presence of other hydrocarbons is reported earlier [55]. In this research, Pseudomonas aeruginosa was identified as potential diesel degrader with maximum degradation of 34.4% at the end of 10 days incubation period. However, with increasing incubation time, the rate of degradation was reduced which could be possible due to the release of by products would inhibit the normal bacterial growth to dramatically reduce the hydrocarbon degradation rate.
Conclusion
The PAH degrading bacteria showed diverse capacities to degrade petrol and diesel and this study is an important step towards the development of bioremediation strategies for cleaning of sites contaminated with polycyclic aromatic hydrocarbons. Further studies are needed to define their bioremediation potential and elucidate the metabolic pathways involved in Petrol and diesel degradation to establish the safety of the by-products from PAH metabolism.
References
- Atlas RM, Hazen TC (1999) Oil biodegradation and bioremediation: A tale of the two worst spills in US history. Environ Sci Technol 45: 6709-6715.
- Liebeg EW, Cutright TJ (1999) The investigation of enhanced bioremediation through the addition of macro and micro nutrients in a PAH contaminated soil. Int Biodeterior Biodegrad 44: 55-64.
- Plohl K, Leskovsek H, Bricelj M (2002) Biological degradation of motor oil in water. Acta Chim Slovenica 49: 279-289.
- Abed MMR, Safi NMD, Koster J, deBeer D, El-Nahhal Y, et al. (2002) Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds. Appl Environ Microbiol 68: 1674-1683.
- Boonchan S, Britz ML, Stanley GA (2000) Degradation and mineralisation of high-molecular weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl Environ Microbiol 66: 10.
- Samanta KS, Singh OV, Jain RK (2002) Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol 20: 243-248.
- Heitkamp M, Franklin W, Cerniglia C (1988) Microbial metabolism of polycyclic aromatic compounds: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol 54: 2549-2555.
- Aislabie J, McLeod M, Fraser R (1998) Potential for biodegradation of hydrocarbons in soil from the Ross Dependency, Antarctica. Appl Microbiol Biotechnol 49: 210-214.
- Zhang Q, Wang D, Li M, Xiang WN, Achal V (2014) Isolation and characterization of diesel degrading bacteria, Sphingomonas sp. and Acinetobacter junii from petroleum contaminated soil. Front Earth Sci 8: 58-63.
- Walworth JL, Woolard CR, Harris KC (2003) Nutrient amendments for contaminated per-glacial soils: Use of cod bone meals as controlled release nutrient source. Cold Reg Sci Technol 37: 81-88.
- Delille D, Pelletier E, Coulon F (2007) The influence of temperature on bacterial assemblages during bioremediation of a diesel fuel contaminated subAntarctic soil. Cold Reg Sci Technol 48: 74-83.
- Minai-Tehrani D, Minoui S, Herfatmanesh AB (2009) Effect of salinity on biodegradation of polycyclic aromatic hydrocarbons (PAHs) of heavy crude oil in soil. Environ. Contam Tox 82: 179-184.
- Zhang X, Li J, Huang Y, Thring R (2010) Surfactant Enhanced Biodegradation of Petroleum Hydrocarbons in Oil Refinery Tank Bottom Sludge. J Can Petrol Technol 49: 34-39.
- Yakubu MB (2007) Biological approach to oil spills remediation in the soil. Afr J Biotechnol 6: 2735-2739.
- Pothuluri J, Cerniglia C (1994) Microbial metabolism of polycyclic aromatic hydrocarbons. In: Chaudry GR (edn.) Biological degradation and bioremediation toxic chemicals. London: Chapman and Hall, pp. 92-124.
- Mehrashi MR, Haghighi B, Shariat M, Naseri S, Naddafi K (2003) Biodegradation of petroleum hydrocarbons in soil. Iranian J Public Health 32: 28-32.
- Chaillan F, A Le Fleche, E Bury, Y Phantavong, P Grimont, et al., (2004) Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res Microbiol 155: 587-595.
- Taki H, Syutsubo K, Mattison GR, Haryana S (2007) Identification and characterization of o-xylene-degrading Rhodoccus sp. Where were dominant species in the remediation of oxylene contaminated soils. Biodegradation 18: 17-26.
- Obayori OS, Ilori MO, Adebusoye SA, Amund OO, Oyetibo GO (2008) Microbial population changes in tropical agricultural soil experimentally contaminated with crude petroleum. Afr J Biotechnol 7: 4512-4520.
- Ahn Y, Sanseverino J, Sayler GS (1999) Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegradation 10: 149-157.
- Ting ASY, Tan CHC, AwCS (2009) Hydrocarbon-degradation by isolate Pseudomonas lundensis UTAR FPE2. Malaysian J Microbiol 5: 104-108.
- Johnsen AR, Wick LY, Harms H (2005) Principles of microbial PAH-degradation in soil. Environ Pollut 133: 71-84.
- Chang R, (1998) Chemistry (6thEdn.), McGraw Hill Company Inc, pp. 962-963.
- Marquez-Rocha FJ, Hernandez-Rodriguez V, Lamela MT (2001) Biodegradation of diesel oil in soil by a microbial consortium. Water Air Soil Pollut 128: 313-320.
- Ganesh A, Lin J (2009) Diesel degradation and biosurfactant production by Gram-positive isolates. Afr J Biotechnol 8: 5847-5854.
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792-1797.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465-W469.
- Cerniglia CE (1992) Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3: 351-368.
- Cheung PY, Kinkle BK (2001) Mycobacterium diversity and pyrene mineralization in petroleum contaminated soils. Appl Environ Microbiol, 67: 2222-2229.
- Chaineau CH, YepremianC, VidalieJF, DucreuxJ, BalleriniD (2003) Bioremediation of a crude oil-polluted soil: biodegradation, leaching and toxicity assessments. Water Air Soil Poll 144: 419-440.
- Wongsa P, Tanaka M, Ueno A, Hasanuzzaman M, Yumoto I (2004) Isolation and characterization of novel strains of Pseudomonas aeruginosa and Serratia marcescens possessing high efficiency to degrade gasoline, kerosene, diesel oil, and lubricating oil. Curr Microbiol 49: 415-422.
- Das K, MukherjeeAK (2007) Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Biores Technol 98: 1339-1345.
- Danne LL, HarjonoI, ZylstraGJ, Haggblom MM(2001) Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl Environ Microbiol 67: 2683-2691.
- Bundy JG, PatonGI, CampbellCD (2004) Combined microbial community level and single species biosensor responses to monitor recovery of oil polluted soil. Soil Biol Biochem 36: 1149-1159.
- Demnerova K, Mackova M, Spevakova, V, Beranova K, Kocha L, et al. (2005) Two approaches to biological decontamination of ground-water and soil polluted by aromatics characterization of microbial populations. Intern Microbiol 8: 205-211.
- Harwood CS, Gibson J (1988) Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 54: 712-717.
- Riser-Roberts E (1992) Bioremediation of petroleum contaminated sites. Boca Raton (FL): CRC Press Inc
- Delille D, Delille B, Pelletier E (2002) Effectiveness of bioremediation of crude oil contaminated Subantarctic Intertidal Sediment: the microbial response. Microb Ecol 44: 118-126.
- Lyle G, Whyte L, Charles WG (1997) Biodegradation of petroleum hydrocarbons by psychrotrophic pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol 63: 3719-3723.
- Leahy JG, Colwell RR (1990) Microbial degradation of hydrocarbons in the environment. Microbiol Rev 54: 305-315.
- Shukor MY, Hassan NA, Jusoh AZ, Perumal N, Shamaan NA (2009) Isolation and characterization of a Pseudomonas diesel-degrading strain from Antarctica. J Environ Biol 30: 1-6.
- Lee M, Kim MK, Singleton I, Goodfellow M, Lee ST (2006) Enhanced biodegradation of diesel oil by a newly identified Rhodococcus baikonurensis EN3 in the presence of mycolic acid. J Appl Microbiol 100: 325-333.
- Mukherji S, Jagadevan S, Mohapatra G, Vijay A (2004) Biodegradation of diesel oil by an Arabian Sea sediment culture isolated from the vicinity of an oil field. Biores Technol 95: 281-286.
- Bicca FC, FleckLC, AntonioM and AyubZ (1999) Production of biosurfactant by hydrocarbon degrading Rhodococcus ruber and Rhodococcus erythropolis. Rev de Microbiol 30: 231-236.
- Espeche ME, MacCormackWP, FraileER (1994) Factors affecting growth of an n- hexadecane degrader Acinetobacter species isolated from a highly polluted urban river. Int Biodeterior Biodegrad 33: 187-196.
- Deng MC, LiJ, FR Liang, M Yi, XM Xu, et al. (2014) Isolation and characterization of a novel hydrocarbon degrading bacterium Achromobacter sp HZ01 from the crude oil contaminated seawater at the Daya Bay, Southern China. Mar Pollut Bull 83: 79-86.
- Dave BP, GhevariyaCM, BhattJK, DudhagaraDR, RajparaRK (2014) Enhanced biodegradation of total polycyclic aromatic hydrocarbons (TPAHs) by marine halotolerant Achromobacter xylosoxidans using Triton X-100 and ß-cyclodextrin--a microcosm approach. Mar Pollut Bull 79: 123-129.
- Tanase AM, Ionescu R, Chiciudean L, Vassu T, StoicaI (2013) Characterization of hydrocarbon-degrading bacterial strains isolated from oil-polluted soil. Int Biodeter Biodegr 84: 150-154.
- Farajzadeh Z, Karbalaei-HeidariHR (2012) Isolation and characterization of a new Achromobacter sp. strain CAR1389 as a carbazole-degrading bacterium. World J Microbiol Biotechnol 28: 3075-3080.
- Mnif S, Chamkha M, Labat M, Sayadi S (2011) Simultaneous hydrocarbon biodegradation and biosurfactant production by oilfield-selected bacteria. J Appl Microbiol 111: 525-536.
- Suen WC, Gibson DT (1993) Isolation and preliminary characterisation of the subunits of the terminal component of naphtalene dioxygenase from Pseudomonas putida NCIB 9816-4. J Bacteriol 175: 5877-5881.
- Mittal A, Singh P (2009) Isolation of hydrocarbon degrading bacteria from soils contaminated with crude oil spills. Indian J Exp Biol 47: 760-765.
- Ueno A, Hasanuzzaman M, Yumoto I, Okuvama H (2006) Verification of degradation of n-alkanes in diesel oil by Pseudomonas aeruginosa strain WatG in soil microcosms. Curr Microbiol 52: 182-185.
- Gallego JL, Loredo J, Llamas JF, Vazquez F, Sanchez J (2001) Bioremediation of diesel-contaminated soils: evaluation of potential in situ techniques by study of bacterial degradation. Biodegradation 12: 325-335.
- Hong J, Kim J, Choi O, Cho KS, H Ryu (2005) Characterization of a diesel-degrading bacterium, Pseudomonas aeruginosa IU5, isolated from oil-contaminated soil in Korea. World J Microbiol Biotechnol 21: 381-384.
Copyright: © 2015 Sibi G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.