Journal of Oceanography and Marine Research
Open Access
ISSN: 2572-3103
ISSN: 2572-3103
Research Article - (2013) Volume 1, Issue 1
The green macroalga Ulva rigida C. Agardh (Chlorophyta) and the red macroalga Gelidium microdon Kützing (Rhodophyta), collected from the Azorean archipelago, were investigated for their secondary metabolites and their in vitro growth inhibitory effect on three human tumor cell lines: MCF-7 (breast adenocarcinoma), NCI-H460 (non-small cell lung cancer) and A375-C5 (melanoma), as well as for their antifungal and antibacterial activities. The methanol extract of U. rigida furnished isofucosterol (1), 7(E)-3β-hydroxy-5α,6α-epoxymegastigmane (2) and (+)-dehydrovomifoliol (3) while the methanol extract of G. microdon yielded cholesterol (4) and lumichrome (5). The crude extracts of both macroalgae were found to be moderately active against the three cell lines whereas compound 1 showed a weak effect and compound 2 was inactive. The crude extracts of the two macroalgae were found to be moderately active against some fungi and bacteria while compounds 1 and 2 were inactive against all microorganisms tested.
<Keywords: Azores; Macroalgae; Ulva rigida; Gelidium microdon; Isofucosterol; 7(E)-3β-hydroxy-5, 6-epoxymegastigmane; (+)-dehydrovomifoliol; Lumichrome; Antitumor; Antimicrobial
MeOH - Methanol; Me2CO - Acetone; δ - Chemical Shift in ppm; DMSO - Dimethyl Sulphoxide; HR-ESIMS-High Resolution Electrospray Ionization Mass Spectrometry; SRB - Sulforhodamine B; MIC - Minimal Inhibitory Concentration; MLC - Minimal Lethal Concentration
The marine environment is an exceptional reservoir of bioactive compounds, many of which exhibit structural/chemical features not found in terrestrial natural products. This is easily understood since the Ocean, which covers almost 71% of the Earth’s surface and represents a uniqueness of the physical and chemical conditions, is considered as a very promising source of Natural Products covering a wide range of bioactivities [1-6]. Therefore, marine Natural Products continue to play a major role in drug discovery.
Since the Azorean archipelago is located in the warm temperate region of the North East Atlantic, approximately 1200 km from Europe, the marine fauna and flora of this group of islands appear to be a mixture of species which can be found both in the Atlantic and the Mediterranean [7]. Marine macroalgae are abundant and structuring organisms of the coastal area of the entire Azores archipelago, some having a markedly seasonal pattern and others being present during the whole year in the Azorean coasts [8-10]. The geographical distribution of these macroalgae is related to the temperature regime of the region where they grow, reproduce, and survive. However, the diversity and abundance of these organisms depend on many other biological factors [11], leading to production of different secondary metabolites of the species from different geographical locations [12]. Macroalgae produce myriads of secondary metabolites which are synthesized at the end of the growth phase and/or due to metabolic alterations induced by environmental stress conditions [13]. These metabolites have been targets of the drug discovery program and some of these bioactive compounds such as sulfated polysaccharides, steroids and diterpenes have found their applications in the pharmaceutical industry [14,15].
During our on-going project aiming at exploiting bioactive secondary metabolites from macroalgae of the Azorean archipelago for added-value products, we have conducted phytochemical studies of the green alga Ulva rigida C. Agardh and the red alga Gelidium microdon Kützing, and evaluation of the in vitro antitumor and antimicrobial activities of the crude extracts of these two macroalgae as well as their isolated metabolites. The main reasons for selection of these two species were based on the fact that Ulva and Gelidium species are well-recognized sources of industrially important biopolymers and the organic crude extracts of these two species had been previously found to exhibit a promising in vitro cytotoxicity on cancer cell lines and antioxidant activity [16]. Furthermore, they are abundant in the Azorean intertidal areas [8,17]. Although both species are locally abundant and dominant at the Azorean intertidal bedrock areas, U. rigida is common and abundant at mid and low shore levels whereas G. microdon is extremely abundant at mid shore level. Consequently, their abundance and easy access for collection can guarantee their quantity for further biotechnological exploitation in the future. Furthermore, as these two species are annual and intertidal, they do not present any significant variations of the concentrations of their secondary metabolites, which can be influenced by their age and depth of the collection site. Although both Ulva and Gelidium species have been extensively investigated as sources of biotechnologically relevant biopolymers, their secondary metabolites have never been fully exploited for value-added products. While Ulva species are an important source of ulvan, a natural sulfated polysaccharide which has been extensively investigated for development of novel drugs and functional foods [18], Gelidium species are one of the main sources of phycocolloids, such as agar [19,20]. Several types of secondary metabolites such as bromophenol [21-23], sesquiterpenes [24,25], and steroids [23,26] have been previously reported from the macroalgae of the genus Ulva; however, there are only few reports on the chemical constituents of the genus Gelidium. While gelidene, a polyhalogenated monocyclic monoterpene, was isolated from G. sesquipedale [27], jasmonic acid was reported from G. latifolium [28].
Due to the pristine environment of the Azorean archipelago, we have elaborated the project aiming to exploit the potential of the macroalgae of this region. The collections of these two species were carried out in May and October in order to allow us to study their chemical compositions in different seasons, i.e. spring and autumn, as well as of two different reproductive stages. We now report the chemical study together with the antitumor and antimicrobial activities evaluation of the first collection (May 2011) of the green macroalga U. rigida and the red macroalga G. microdon from S. Miguel Island which is considered to be one of the environmentally healthy habitats and rich in algal communities of the Azorean Sea. Examination of the methanol extract of U. rigida led to isolation of isofucosterol (1), 7(E)- 3β-hydroxy-5α,6α-epoxymegastigmane (2) and (+)-dehydrovomifoliol (3), while the methanol extract of G. microdon yielded cholesterol (4) and lumichrome (5) (Figure 1). The crude extracts of both macroalgae, together with isofucosterol (1) and 7(E)-3β-hydroxy- 5α,6α-epoxymegastigmane (2), were evaluated for their in vitro growth inhibition on three tumor cell lines: MCF-7, NCI-H460 and A375-C5, as well as for their antifungal and antibacterial activities. Figure 1.
General experimental procedures
Melting points were determined on a Bock monoscope and are uncorrected. Optical rotations were determined on an ADP410 Polarimeter. 1H and 13C NMR spectra were recorded at ambient temperature on a Bruker Advance instrument operating at 300.13 and 75.4 MHz, respectively. High resolution mass spectra were measured with a Waters Xevo QToF mass spectrometer coupled to a Waters Aquity UPLC system. A Merck silica gel GF 254 was used for preparative TLC, and a Merck Si gel 60 (0.2-0.5 mm) was used for analytical chromatography.
Biological material
U. rigida and G. microdon were collected in May 2011 from the coast of S. Miguel island - Azores archipelago, and the samples of both macroalgae were deposited at the Department of Biology of University of Azores (Vouchers: SMG-11-49 and SMG-11-30, respectively).
Extraction and isolation of the constituents
Dried powdered material (U. rigida - 1427.5 g and G. microdon - 1293.4 g) was percolated with MeOH, at room temperature until exhaustion. The resulting solutions were filtered with filter paper (Whatman no 1) and concentrated under reduced pressure to yield crude extracts of U. rigida (154.49 g) and G. microdon (151.91 g). Treatment of the crude methanol extracts to remove the chlorophylls [29], furnished 14.22 g of U. rigida and 4.74 g of G. microdon purified extracts
U. rigida - The purified extract (14.22 g) was chromatographed over a 0.2-0.5 mm Si Gel column (180 g) and eluted with mixtures of petroleum ether, CHCl3 and Me2CO, wherein 250 ml fractions were collected as follows: frs 1-2 (petroleum ether-CHCl3, 9:1), frs 3-52 (petroleum ether/CHCl3, 4:1), frs 53-112 (petroleum ether-CHCl3, 1:1), frs 113-126 (petroleum ether-CHCl3, 1:4), frs 127-145 (CHCl3), frs 146- 167 (CHCl3-Me2CO, 9:1), frs 168-211 (CHCl3-Me2CO, 4:1), frs 212-243 (CHCl3-Me2CO, 1:1), frs 244-289 (CHCl3-Me2CO, 1:4), frs 290-303 (Me2CO). Frs 30 and 31 were combined (691.1 mg) and recrystallized in petroleum ether to give 32.0 mg of compound 1. Frs 93-106 were combined (121.9 mg) and purified by TLC (Si Gel, CHCl3-EtOAcpetroleum ether-HCO2H; 3:1:1:0.01), to yield compound 2 (10.8 mg) and a mixture (4.6 mg) containing compound 3 as a major component.
G. microdon - The purified extract (4.74 g) was chromatographed over a 0.2-0.5 mm Si Gel column (120 g) and eluted with mixtures of petroleum ether, CHCl3, Me2CO and MeOH, 250 ml fractions were collected as follows: frs 1-23 (petroleum ether-CHCl3, 4:1), frs 24-73 (petroleum ether-CHCl3, 3:2), frs 74-112 (petroleum ether-CHCl3, 1:4), frs 113-125 (petroleum ether-CHCl3, 1:9), frs 126-142 (CHCl3), frs 143-175 (CHCl3-Me2CO, 9:1), frs 176-200 (CHCl3-Me2CO, 7:3), frs 201-231 (CHCl3-Me2CO, 1:1), frs 232-236 (CHCl3-Me2CO, 3:7), frs 237-256 (CHCl3-MeOH, 9:1), frs 257-267 (CHCl3-MeOH, 4:1), frs 268- 280 (CHCl3-MeOH, 1:1), frs 281-294 (CHCl3-MeOH, 3:7), frs 295-300 (MeOH). Frs 11-22 were combined (665.4 mg) and purified by TLC (Si gel, CHCl3-toluene-HCO2H, 7:3:0.1) to give 66.9 mg of cholesterol (4) [30]. Frs 81-100 were combined (127.6 mg) and recrystallized in CHCl3 to yield 2.7 mg of lumichrome (5).
Isofucosterol (1): White crystals; mp. 129-131°C. 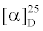 = - 36.2° (c 0.083 g mL-1, CHCl3) Lit: mp 128-130°C.
= - 36.2° (c 0.083 g mL-1, CHCl3) Lit: mp 128-130°C. 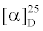 = - 36.8° (c 0.02 g mL-1, CHCl3) [31]. 1H- NMR (300 MHz, CDCl3) δ in ppm: 5.35 dd (J=1.7, 4.4 Hz, H-6), 5.10 q (J=6.8 Hz, H-28), 3.52 dddd(J=1.2, 5.6, 10.7, 10.7 Hz, H-3), 2.83 m (H-25), 2.29 m (H-4), 1.9-2.1 m (H-7, H-8, H-12, H-23), 1.7-1.9 m (H-1, H-2, H-16, H-23), 1.4-1.6 m (H-7, H-8, H-11, H-15, H-20, H-22), 0.9-1.3 m (H-9, H-12, H-14, H-15, H-16, H-17, H-22), 1.59 d(J=6.8 Hz, H-29), 1.01 s (H-19), 0.95 d(J=6.6 Hz, H-21), 0.98 d(J=6.9 Hz, H-26, H-27), 0.68 s (H-18). 13C-NMR (75 MHz, CDCl3) δ in ppm: 145.9 s(C-24), 140.7 s(C-5), 121.7 d(C-6), 116.4 d(C-28), 42.3 s(C- 13), 42.3 t(C-4),71.8 d(C-3), 56.7 d(C-14), 56.0 d(C-17), 50.1 d(C-9), 39.8 t(C-12), 37.2 t(C-1), 36.5 s(C-10), 36.1 (C-20), 35.9 (C-22), 32.0 t(C-7), 31.9 d(C-8), 31.6 t(C-2), 28.6 d(C-25), 27.9 t(C-23), 28.2 t (C-16), 24.3 t(C-15), 21.2 t(C-11), 21.1 q(C-26), 21.0 q(C-27), 19.4 q (C-19), 18.8 q(C-21), 11.8 q(C-18), 12.8 q(C-29).
= - 36.8° (c 0.02 g mL-1, CHCl3) [31]. 1H- NMR (300 MHz, CDCl3) δ in ppm: 5.35 dd (J=1.7, 4.4 Hz, H-6), 5.10 q (J=6.8 Hz, H-28), 3.52 dddd(J=1.2, 5.6, 10.7, 10.7 Hz, H-3), 2.83 m (H-25), 2.29 m (H-4), 1.9-2.1 m (H-7, H-8, H-12, H-23), 1.7-1.9 m (H-1, H-2, H-16, H-23), 1.4-1.6 m (H-7, H-8, H-11, H-15, H-20, H-22), 0.9-1.3 m (H-9, H-12, H-14, H-15, H-16, H-17, H-22), 1.59 d(J=6.8 Hz, H-29), 1.01 s (H-19), 0.95 d(J=6.6 Hz, H-21), 0.98 d(J=6.9 Hz, H-26, H-27), 0.68 s (H-18). 13C-NMR (75 MHz, CDCl3) δ in ppm: 145.9 s(C-24), 140.7 s(C-5), 121.7 d(C-6), 116.4 d(C-28), 42.3 s(C- 13), 42.3 t(C-4),71.8 d(C-3), 56.7 d(C-14), 56.0 d(C-17), 50.1 d(C-9), 39.8 t(C-12), 37.2 t(C-1), 36.5 s(C-10), 36.1 (C-20), 35.9 (C-22), 32.0 t(C-7), 31.9 d(C-8), 31.6 t(C-2), 28.6 d(C-25), 27.9 t(C-23), 28.2 t (C-16), 24.3 t(C-15), 21.2 t(C-11), 21.1 q(C-26), 21.0 q(C-27), 19.4 q (C-19), 18.8 q(C-21), 11.8 q(C-18), 12.8 q(C-29).
7(E)-3β-Hydroxy-5α, 6α-epoxymegastigmane (2): Oil; 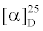 = - 38.5° (c 0.026 g mL-1, CHCl3) Lit:
= - 38.5° (c 0.026 g mL-1, CHCl3) Lit: 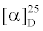 = -43.7° (c 0.39 g mL-1, CH2Cl2) [32]. 1H and 13C NMR (Table 1).
= -43.7° (c 0.39 g mL-1, CH2Cl2) [32]. 1H and 13C NMR (Table 1).
| Position | δC, type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 35.1, C | |||
| 2α | 46.6, CH2 | 1.64, dt (12.9, 1.8) | H-2β | |
| β | 1.26, dt (12.9, 10.4) | H-2α | ||
| 3 | 64.0, CH | 3.90, m | ||
| 4α | 40.5, CH2 | 2.39, ddd (14.5, 5.1, 1.7) | H-2β | C-3, 5, 6 |
| β | 1.66, dd (14.5, 8.7) | H-2α | C-2, 5 | |
| 5 | 67.3, C | |||
| 6 | 69.4, C | |||
| 7 | 142.4, CH | 7.03, d (15.6) | H-8 | C-6, 8, 9 |
| 8 | 132.6, CH | 6.29, d (15.6) | H-7 | C-6, 7, 9 |
| 9 | 197.5, CO | |||
| 10 | 28.3, CH3 | 2.28, s | ||
| 11 | 19.8, CH3 | 1.19, s | C-4, 6 | |
| 12 | 25.0, CH3 | 0.98, s | C-1, 2, 6, 13 | |
| 13 | 29.3, CH3 | 1.19, s | C-1, 2, 6, 12 |
Table 1: NMR data for compound 2 in CDCl3 (1H 300.13, 13C 75.47 MHz).
(+)-Dehydrovomifoliol (3): White amorphous powder. 1H and 13C NMR (Table 2).
| Position | δC, type | δH, (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 41.4, C | |||
| 2α | 49.5, CH2 | 2.51, d (17.0) | H-2 β | C-1, 3, 11 |
| β | 2.34, d (17.0) | H-2 α | ||
| 3 | 197.0, CO | |||
| 4 | 127.8, CH | 5.96, brt (1.0) | H-2 β, 13 | C-2, 6, 13 |
| 5 | 160.4, C | |||
| 6 | 79.3, C | |||
| 7 | 145.0, CH | 6.84, d (15.7) | H-8 | C-5, 6, 9 |
| 8 | 130.3, CH | 6.47, d (15.7) | H-7 | C-6, 9 |
| 9 | 197.4, CO | |||
| 10 | 28.4, CH3 | 2.31, s | C-8, 9 | |
| 11 | 18.7, CH3 | 1.88, s | C-4, 5, 6 | |
| 12 | 24.3, CH3 | 1.03, s | C-1, 2, 13 | |
| 13 | 22.9, CH3 | 1.11, s | C-1, 2, 12 |
Table 2: NMR data for compound 3 in CDCl3 (1H 300.13, 13C 75.47 MHz).
Cholesterol (4): White crystals; mp. 146-147°C; 1H NMR (300 MHz, CDCl3) δ 5.36 d(J=5.2 Hz, H-5), 3.53 m (H-3), 2.2-2.4 m (H-4 and H-13), 1.9-2.1 m (H-7, H-12) 1.4-1.6 m (H-1 and H-2), 1.02 s (H- 19, 3H), 0.92 d(J=6.5 Hz, H-21, 3H), 0.88 d(J=6.6 Hz, H-27, 3H), 0.87 d(J=6.6 Hz, H-27, 3H), 0.69 s (H-18, 3H); 13C-NMR (75 MHz, CDCl3) δ 140.7 s(C-5), 121.7 d(C-6), 71.8 d(C-3), 56.8 d(C-17), 56.1 s(C-14), 50.1 d(C-9), 42.3 t(C-4 and C-13), 39.8 t(C-12), 39.5 t(C-24), 37.2 t (C-1), 36.5 s(C-10), 36.2 t(C-22), 35.8 d(C-20), 31.9 t(C-7), 31.9 t(C- 8), 31.7 t(C-2), 28.2 t(C-16), 28.0 d(C-25) 24.3 t(C-15), 23.8 t(C-23), 22,8 q(C-27), 22.6 q(C-26), 21.1 t(C-11), 19.4 q(C-19), 18.7 q(C-21), 11.9 q(C-18).
Lumichrome (5): Green amorphous powder. HR-ESIMS m/z 243.0918 [M+H]+ (calcd for C12H11N4O2 , 243.0882); 1H and 13C NMR (Table 3).
| Position | δC, type | δH, (J in Hz) | HMBC |
|---|---|---|---|
| 1 | |||
| 2 | 130.2, C | ||
| 3 | 146.5, C | ||
| 4 | |||
| 5 | 138.4, C | ||
| 6 | 128.7, CH | 7.92, s | C-7, 13, 15 |
| 7 | 138.9, C | ||
| 8 | 144.7, C | ||
| 9 | 125.8, CH | 7.71, s | C-6, 12, 16 |
| 10 | 141.6, C | ||
| 11 | 11.84, brs | C-2 | |
| 12 | 150.1, CO | ||
| 13 | 11.68, brs | C-2 | |
| 14 | 160.7, CO | ||
| 15 | 19.6, CH3 | 2.47, s | C-11, 12, 13 |
| 16 | 20.2, CH3 | 2.49, s | C-9, 12, 13 |
Table 3: NMR data for compound 5 in CDCl3 (1H 300.13, 13C 75.47 MHz).
Growth inhibition of human tumor cell lines
The effect of the extracts and of compounds 1 and 2 were evaluated for their capacity to inhibit in vitro growth of three human tumor cell lines: MCF-7 (breast adenocarcinoma), NCI-H460 (non-small cell lung cancer) and A375-C5 (melanoma), according to the procedure adopted by the National Cancer Institute (NCI) in the “In vitro Anticancer Drug Discovery Screen” that uses the protein-binding dye SRB to assess cell growth as previously described [33]. The cell cultures used were from different sources. MCF-7 and A375-C5 were obtained from the European Collection of Cell Cultures (ECACC) while NCI-H460 was provided by the NCI collection of cell lines. Briefly, compounds 1, 2 and the crude extracts were aseptically dissolved in DMSO and stored at -20°C. Appropriate dilutions of the compounds and extracts were freshly prepared just prior the assay and diluted with growth medium. Final concentrations of DMSO did not interfere with the growth of cell lines. Cells growing as monolayer, were routinely maintained in RPMI-1640 medium containing 5% FBS, at 37°C in a humidified atmosphere containing 5% CO2. For the assays, each cell line was plated at an appropriate density (5x104 cells/mL) in 96 well-plates. Cells were incubated for 24h in the humidified incubator, allowing them to stabilize and adhere. Cells were then exposed for 48h at 37°C and 5% CO2 to serial concentrations of compounds, extracts and the positive control, doxorubicin. Following this exposure period, cells were fixed in situ with 50% Trichloroacetic Acid (TCA), washed with distillated water and dyed with SRB. In order to solubilize protein/SRB complexes, Tris buffer were added to each well and absorbance (Abs at 515 nm) was determined in a plate reader (Biotek Synergy 2). Abs values were retrieved using Gene5software (Biotek). For each cell line a dose-response curve was obtained and the growth inhibition of 50% (GI50) corresponding to the concentration of compounds and extracts that inhibited 50% of the net cell growth was calculated according to the procedure adopted by the NCI.
Antifungal assays
Broth microdilution methods based on Clinical and Laboratory Standards Institute (CLSI) reference protocols M27-A3 and M38-A2 for yeasts(Candida albicans) and filamentous fungi (Aspergillus fumigatus and dermatophytes), respectively, were used to determine the MIC and MLC of the crude extracts and the isolated metabolites [34]. Candida albicans ATCC 10231, Aspergillus fumigatus ATCC 46645 and dermatophytes: Epidermophyton floccosum FF9, Microsporum canis FF1, Microsporum gypseum FF3, Trichophyton mentagrophytes FF7, and Trichophyton rubrum FF5 were used as test organisms. The yeast cell suspensions were prepared in 0.85% NaCl and the transmittance of cell density adjusted to that produced by a 0.5 McFarland standard for C. albicans. To achieve an inoculum size of 1-5×103 CFU/mL for C. albicans, the cell suspension was diluted with RPMI 1640. For filamentous fungi, the spore suspensions were prepared in 0.85% NaCl with Tween 20 and the cell density adjusted at 20-250 conidia/ square (hemocytometer) for A. fumigatus and 20-60 conidia/square for dermatophytes. To achieve an inoculum size of 0.4-5×104CFU/mL for A. fumigatus and 1-3×103 CFU/mL for dermatophytes, the spore suspensions were diluted with RPMI 1640. The solutions of the extracts and compounds 1 and 2 were prepared in DMSO and added to the cell suspensions in order to obtain test concentrations ranging from 16 to 256 μg/mL. In addition, reference antifungal compound, fluconazole was used as standard antifungal drug. Controls without crude extracts and isolated compounds, as well as sterility and DMSO control wells, were also included. The plates were incubated aerobically at 35°C ± 0.2°C for 24h/48h in atmospheric humidity (C. albicans and A. fumigatus) and at 25°C ± 0.2°C for 5 days in atmospheric humidity for dermatophytes. To evaluate the MLCs, 20 μL samples were taken from each negative well and the first well exhibiting growth (serve as a growth control), after MIC reading, spotted onto SDA (Sabouraud Dextrose Agar) plates and incubated at 35°C ± 0.2°C 24h/48h (C. albicans and A. fumigatus) or at 25°C ± 0.2°C for 7 days (dermatophytes).
Antibacterial assays
A broth microdilution method, based on CLSI reference protocol M7-A7, was used to determine the MIC and MLC of the crude extracts and the isolated metabolites [35]. Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923, and Methicillin Resistant Staphylococcus aureus (MRSA), clinical isolate, were used as test organisms. The cell suspensions were prepared in 0.85% NaCl and the transmittance of cell density adjusted to that produced by a 0.5 McFarland standard. To achieve an inoculum size of 105 CFU/mL, the cell suspensions were diluted with MHB (Muller-Hinton Broth). The stock solutions of the extracts, and compounds 1 and 2 were prepared in DMSO and further diluted in serial two-folds with MHB to final concentrations ranging from 16 to 256 μg/mL. In addition, gentamicin was used as standard antibacterial drug and controls without crude extracts and isolated compounds, as well as sterility and DMSO control wells, were also included. The plates were incubated aerobically at 35°C ± 0.2°C for 16h/20h in atmospheric humidity. To measure the MLCs, 20 μL samples were taken from each negative well and the first well exhibiting growth (serve as a growth control), after MIC reading, spotted onto MHA (Muller-Hinton Agar) plates and incubated at 35°C ± 0.2°C for 24h.
The structures of the compounds were established mainly by 1D (1H and 13C NMR) and 2D (COSY, DEPT, HSQC and HMBC experiments) spectroscopic techniques as well as comparison of their NMR data with those reported in the literature.
Compound 1 was isolated as white crystals with mp 129-131°C. The 13C NMR, DEPT and HSQC spectra revealed the presence of twenty nine carbon signals which can be categorized as two quaternary sp2 (δc 145.9, 140.7), two methine sp2 (δc 121.7, 116.4), two quaternary sp3 (δc 42.3, 36.5), one oxymethine sp3 (δc 71.8), six methine sp3 (δc 56.7, 56.0, 50.1, 36.1, 31.9, 28.6), ten methylene sp3 (δc 42.3, 39.8, 37.2, 35.9, 31.9, 31.6, 28.2, 27.9, 24.3, 21.1) and six methyl (δc 21.1, 21.0, 19.4, 18.8, 12.8, 11.8) carbons. The HMBC correlations of H-6 (δH 5.35, dd, J= 4.4, 1.7, δc 121.7) to C-4 (δc 42.3), C-8 (δc 31.9), C-10 (δc 36.5) revealed the presence of a trisubstituted double bond between C-5 and C-6. That another trisubstituted double bond was on C-24 and C-28 was corroborated by the HMBC correlations of H-28 (δH 5.10, q, J = 6.8, δc 116.4) to C-25 (δc 28.6) and C-29 (δc 12.8). The coupling constants of H-3 suggested that the C-3 hydroxyl group was β. The 1H and 13C chemical shift values of compound 1 were compatible with those of isofucosterol [36,37]. Isofucosterol is a common phycosterol and it has been previously reported from several macroalgae [38].
The 13C NMR spectrum of compound 2 displayed thirteen carbon signals which were categorized, by DEPT and HSQC experiments (Table 1), as one carbonyl of a conjugated ketone (δc 197.5), two methine sp2 (δc 132.6, 142.4), two oxyquaternary sp3 (δc 69.4, 67.3), one quaternary sp3 (δc 35.1), one oxymethine sp3 (δc 64.0), two methylene sp3 (δc 40.5, 46.6) and four methyl (δc 19.8, 24.9, 28.3, 29.3) carbons. The COSY spectrum displayed cross peak between the olefinic protons at δH 7.03 d(J= 15.6) and δH 6.29 d(J= 15.6), confirming the presence of a trans double bond. That this trans double bond was part of the 3-oxobutenyl side chain which linked to C-6 of the cyclohexanol moiety was supported by the HMBC correlations of the methyl protons signal at δH 2.28s (δC 28.3) to the carbon signals at δC 132.6 (C-8), δC 142.4 (C-7) and δC 197.5 (C-9), as well as of the proton signal at δH 6.29d, J = 15.6 (δC 132.6) to the carbon signals at δC 197.5 (C-9) , δC 142.4 (C-7), δC 69.4 (C-6). As the proton signals of the methyl groups at δH 0.98 (δC 25.0) and δH 1.19s (δC 29.3) gave cross peaks to the quaternary carbon signal at δC 35.1 (C-1) as well as to the carbon signals at δC 46.6 (C-2) and δC 69.4 (C-6), they were assigned for C-12 and C-13, respectively. These correlations led to the conclusion that the structure of compound 2 should correspond to 7(E)-3β-hydroxy-5α, 6α-epoxymegastigmane. As compound 2 is levorotatory with 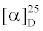 = -38.5°, it was identified as 7(E)-3β-hydroxy-5α, 6α-epoxymegastigmane. The 1H and 13C NMR data of compound 2 were compatible with those of (3S, 5R, 6S, 7E)- 5, 6-epoxy-3-hydroxy-7-megastigmen-9-one, previously isolated from the solanaceous plant Cestrum parqui L’ Hérit [32] .
= -38.5°, it was identified as 7(E)-3β-hydroxy-5α, 6α-epoxymegastigmane. The 1H and 13C NMR data of compound 2 were compatible with those of (3S, 5R, 6S, 7E)- 5, 6-epoxy-3-hydroxy-7-megastigmen-9-one, previously isolated from the solanaceous plant Cestrum parqui L’ Hérit [32] .
Compound 3 was isolated as a mixture, as was evidenced by its 1H NMR spectrum. Through the HMBC correlations, two sets of carbon signals could be discerned in the 13C NMR spectrum of the mixture (Table 2). The first set of the carbon signals, belonging to compound 3, comprised of thirteen carbon signals which were identified by DEPT and HSQC as two carbonyls (δC 197.4 and δC 197.0), one quaternary sp2 (δC 160.4), three methine sp2 (δC 127.8, 130.3, 145.0), one oxyquaternary sp3 (δC 79.3), one quaternary sp3 (δC 41.2), one methylene (δC 49.5), and four methyl (δC 18.7, 22.9, 24.3, 28.4) carbons. Similar to compound 2, there was a 3-oxo-butenyl side chain which linked to the cyclohexenone moiety through the oxyquaternary sp3 carbon at δC 79.3, as was evidenced by the HMBC correlations of the methyl proton signal at δC 2.31s(CH3-10) to C-8 (δC 130.3) and C-9 (δC 197.4), as well as of H-8 signal (δH 6.47, d, J = 15.7 Hz) to C-9 (δC 197.4) and C-6 (δC 79.3). The presence of the 3,5,5-trimethyl-4-hydroxy-2-cyclohexenone moiety was confirmed by the HMBC correlations of the olefinic proton signal at δH 5.96, brt, J = 1.0 Hz (δC 127.8) to CH3-11 (δC 18.7), C-2 (δC 49.5), C-6 (δC 79.3), as well as of the methyl proton signals at δH 1.03s (δH 24.3, CH3-12) and δH 1.11s (δH 22.9, CH3-13) to C-1 (δC 41.2), C-2 (δC 49.5) and C-6 (δC 79.3). The structure of compound 3 was established as (+)-dehydrovomifoliol, previously isolated from several plant sources [39]. However, due to the small quantity of the mixture isolated, it was not possible to isolate 3 as a pure compound to determine its optical rotation .
Compound 5 was isolated as green amorphous powder and its molecular formula C12H10N4O2 was established on the basis of the (+)-HR-ESIMS m/z 243.0918 [M+H]+ (calcd 243.0882), indicating ten degrees of unsaturation. The 13C NMR, DEPT and HSQC spectra (Table 3) revealed the presence of two amide carbonyls (δC 160.7, 150.1), six quaternary sp2 (δC 130.2, 138.4, 138.9, 141.6, 144.7, 146.5), two methine sp2 (δC 125.8, 128.7) and two methyl (δC 19.6, 20.2) carbons. The HMBC spectrum displayed cross peaks of the amide proton signals at δH 11.7 (NH-13) and δH 11.8 (NH-11) to the carbon signal at δC 130.2 (C-2). While the proton signal at δH 7.92s (δC 128.7) showed HMBC correlations to C-8 (δC 144.7), C-10 (δC 141.6) and CH3-15 (δC 19.6), the proton signal at δH 7.71s (δC 125.9) showed HMBC correlations to C-5 (δC 138.4), C-7 (δC 138.9) and CH3-16 (δC 20.2). Thus, the structure of compound 5 is 7, 8-dimethylalloxazine or commonly known as lumichrome. Lumichrome, a derivative of the vitamin riboflavin, has been purified and chemically identified from culture filtrates of the alga Chlamydomonas as a Quorum Sensing (QS) signal-mimic compound capable of stimulating the Pseudomonas aeruginosa LasR QS receptor [40]. Bacteria, plants, and algae commonly secrete riboflavin or lumichrome, raising the possibility that these compounds could serve as either QS signals or as interkingdom signal mimics capable of manipulating QS in bacteria with a LasR-like receptor [40] .
The effect of the extracts of U. rigida and G. microdon (before and after removal of the chlorophylls), isofucosterol (1) and 7(E)- 3β-hydroxy-5α, 6α-epoxymegastigmane (2) were evaluated for their capacity to inhibit in vitro growth of three tumor cell lines: MCF-7, NCI-H460 and A375-C5. The results showed that the crude extracts were moderately active against the three cell lines; however, isofucosterol (1) was found to be less active than the crude extract of U. rigida, while 7(E)-3β-hydroxy-5α,6α-epoxymegastigmane (2) was inactive (Table 4).
| Extract/Compounds | Cell lines / GI50 (µg/ml) | ||
|---|---|---|---|
| MCF-7 | NCI-H460 | A375-C5 | |
| U. rigida(before removal of chlorophylls) | 44.5 ± 18.4 | 49.1 ± 14.0 | 40.8 ± 10.2 |
| U. rigida (after removal of chlorophylls) | 43.0 ± 10.3 | 41.9 ± 12.1 | 44.5 ± 7.6 |
| G. microdon(before removal of chlorophylls) | 75.9 ± 16.1 | 70.6 ± 20.1 | 36.3 ± 8.0 |
| G. microdon(after removal of chlorophylls) | 63.1 ± 14.1 | 64.9 ± 16.6 | 62.6 ± 15.9 |
| Cell lines / GI50 (µM) | |||
| MCF-7 | MCF-7 | MCF-7 | |
| 1 | 122.2 ± 17.9 | 128.4 ± 32.4 | 119.2 ± 28.9 |
| 2 | ≥ 200 | ≥ 200 | ≥ 200 |
Table 4: Growth inhibitory effect of crude methanol extracts of U. rigida and G. microdon, compounds 1 and 2, in different cell lines*.
The crude methanol extracts of U. rigida and G. microdon (before and after removal of the chlorophylls) were also evaluated for their antifungal activity against C. albicans, A. fumigatus, and dermatophytes E. floccosum, M. canis, M. gypseum, T. mentagrophytes, and T. rubrum. The results showed that removal of the chlorophylls caused an increase in antifungal activity of U. rigida against T. rubrum, T. mentagrophytes, M. canis, and E. floccosum. Whereas T. rubrum showed higher susceptibility, M. gypseum showed more resistance (MIC higher than 256 μg/mL). Removal of the chlorophylls also caused an increase in the activity of G. microdon crude extract against T. rubrum and E. floccosum. It was found that M. canis showed more susceptibility while T. mentagrophytes and M. gypseum showed higher resistance. Interestingly, both isofucosterol (1) and 7(E)-3β-hydroxy-5α,6α- epoxymegastigmane (2) were inactive against all the tested organisms (Table 5).
| Extract/ Compound | Fungi / µg /mL | |||||||
|---|---|---|---|---|---|---|---|---|
| C. albicans | A. fumigatus | T. rubrum | T. mentagrophytes | E. floccosum | M. canis | M. gypseum | ||
| U. rigida(before removal of chlorophylls) | MIC | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| U. rigida (after removal of chlorophylls) | MIC | >256 | >256 | 64-128 | 128-256 | 128-256 | 128 | >256 |
| MLC | >256 | >256 | 128 | 128-256 | ≥256 | 128 | >256 | |
| G. microdon (before removal of chlorophylls) | MIC | >256 | >256 | ≥256 | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | ≥256 | >256 | >256 | >256 | >256 | |
| G. microdon (after removal of chlorophylls) | MIC | >256 | >256 | 64-128 | 256 | 64-256 | 64 | >256 |
| MLC | >256 | >256 | 64-128 | ≥256 | 128-256 | 64 | >256 | |
| 1 | MIC | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| 2 | MIC | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
Table 5: Antifungal effect of the crude methanol extracts of U. rigida and G. microdon, compounds 1 and 2, in different selected fungi*.
The extracts of U. rigida and G. microdon (before and after removal of the chlorophylls) were evaluated for their activity against E. coli, P. aeruginosa, S. aureus, and MRSA. The results (Table 6) showed that the crude methanol extract of U. rigida (before and after removal of the chlorophylls) did not show any antibacterial activity against E. coli, P. aeruginosa and S. aureus, however removal of the chlorophylls caused a weak activity against MRSA. Similarly, the crude methanol extract of G. microdon did not show any activity against the test bacteria; however removal of the chlorophylls showed a weak activity against S. aureus and that sensitivity increases against MRSA. Interestingly, neither isofucosterol (1) nor 7(E)-3β-hydroxy-5α,6α-epoxymegastigmane (2) showed activity against all the strains of tested organisms (Table 6).
| Extract/ Compound | Bacteria / µg /mL | ||||
|---|---|---|---|---|---|
| E. coli | P. aeruginosa | S. aureus | MRSA | ||
| U. rigida(before removal of chlorophylls) | MIC | > 256 | > 256 | > 256 | > 256 |
| MLC | > 256 | > 256 | > 256 | > 256 | |
| U. rigida(after removal of chlorophylls) | MIC | > 256 | > 256 | > 256 | 128 |
| MLC | > 256 | > 256 | > 256 | > 256 | |
| G. microdon (before removal of chlorophylls) | MIC | > 256 | > 256 | > 256 | > 256 |
| MLC | > 256 | > 256 | > 256 | > 256 | |
| G. microdon (after removal of chlorophylls) | MIC | > 256 | > 256 | 64 | 32-64 |
| MLC | > 256 | > 256 | > 256 | 64-128 | |
| 1 | MIC | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | >256 | >256 | |
| 2 | MIC | >256 | >256 | >256 | >256 |
| MLC | >256 | >256 | >256 | >256 | |
Table 6: Antibacterial effect of the crude methanol extracts of U. rigida and G. microdon, compounds 1 and 2, in different selected bacteria*.
This work was financially supported by the project “Bioactive products in marine algae of Azores (PTDC/MAR/100482/2008)”, through Fundação para a Ciência e a Tecnologia (FCT), COMPETE, QREN, FEDER, MCTES, and partially supported by CEQUIMED-PEst-OE/SAU/UI4040/2011, Project “PEst-C/MAR/ LA0015/2011 and QOPNA-PEst-C/QUI/UI0062/2011. Madalena Silva thanks FCT for the young researcher scholarship under the PTDC/MAR/100482/2008 project.