Chemotherapy: Open Access
Open Access
ISSN: 2167-7700
ISSN: 2167-7700
Research Article - (2023)Volume 11, Issue 1
Objective: Acute Myeloid Leukemia (AML) is one of the most common aggressive hematological malignancies. circDPY19L1P1 have been demonstrated to present upregulation in AML patients. We attempted to elucidate molecular mechanism of circDPY19L1P1 underlying AML cell malignancy.
Methods: RT-qPCR evaluated circDPY19L1P1 level in AML bone marrow specimens and cells. RNase R digestion assay and actinomycin D evaluated circDPY19L1P1 circular characteristics in AML cells. FISH determined circDPY19L1P1 subcellular distribution in AML cells. Loss-of-function assays clarified circDPY19L1P1 role in AML cell behaviors. Bioinformatics and mechanism experiments assessed association of circDPY19L1P1 or PNPLA6 with miR-130a-3p in AML cells. Rescue assays assessed regulatory function of PNPLA6 in circDPY19L1P1-meidated AML cellular phenotypes.
Results: CircDPY19L1P1 presented upregulation and stable circular characteristics in AML cells. CircDPY19L1P1 silencing suppressed AML cell proliferation facilitated AML cell apoptosis and blocked AML cell differentiation. CircDPY19L1P1 served as a miR-130a-3p sponge to downregulate miR-130a-3p in AML cells. MiR-130a-3p targeted PNPLA6 3'UTR in AML cells and circDPY19L1P1 competitively bound to miR-130a-3p to upregulate PNPLA6. The influences of circDPY19L1P1 knockdown on AML cell proliferative ability, apoptosis and cell differentiation were countervailed by PNPLA6 elevation.
Conclusion: CircDPY19L1P1 presents upregulation and acts as an oncogene in AML cellular behaviors. CircDPY19L1P1 facilitates AML cell malignancy through serving as miR-513a-5p sponge to upregulate PNPLA6, which may provide a potential novel insight for therapeutic strategies of AML.
CircDPY19L1P1; miR-130a-3p; PNPLA6; Acute Myeloid Leukemia (AML); Differentiation
AML: Acute Myeloid Leukemia; ROC: Receiver Operating Characteristic; ITP: Idiopathic Thrombocytopenic Purpura; FISH: Fluorescence In Situ Hybridization
Acute Myeloid Leukemia (AML) is one of the most common aggressive hematological malignancies, characterized by abnormal proliferation and blocked differentiation of immature myeloid cells, accounting for approximately 70%-80% of acute leukemia [1]. Relative to other types of leukemia, mortality of adult AML patients ranks first [2]. According to the characteristics of cell morphology, immune markers, genetics, etc., AML can be classified into different subtypes with high heterogeneity [3]. In recent years, molecular targeted therapy, immunotherapy, hematopoietic stem cell transplantation, etc., for AML have made great advancements; nevertheless, due to recurrence and drug resistance, lots of patients still die of AML progression every year, and overall disease-free survival of AML patient’s remains low [4,5]. Thus, clarifying molecular mechanism underlying AML occurrence and development of AML is of great significance for figurine out new clinical biomarkers and therapeutic targets.
With advancement of high-throughput sequencing technology, circular RNAs (circRNAs) have been demonstrated to present wide distribution in eukaryotes and perform complex biological functions [6]. Relative to messenger RNAs (mRNAs), circRNAs are more stable, can tolerate degradation by RNA enzyme to a certain extent, with high spatial, temporal and tissue specificity [7]. CircRNAs exert crucial biological functions in various benign and malignant diseases, including AML [8,9]. CircVIM presents a marked upregulation in nonacute promyelocytic leukemia patients; through drawing Receiver Operating Characteristic (ROC) curve, circVIM can distinguish nonacute promyelocytic leukemia from AML patients with normal cytogenetics, suggesting that circVIM is an ideal diagnostic marker for AML [10]. Li et al. have demonstrated that hsa_circ_0004277 presents downregulation in AML patients and shows further downregulation in relapsed refractory AML patients, suggesting that hsa_circ_0004277 can be utilized for monitoring early relapse of AML [11]. Ping et al., have validated that circ_0009910 upregulation can facilitate AML cell proliferation, whereas circ_ 0009910 silencing can suppress cell proliferation [12]. The above reports indicate that circRNAs exert vital roles in AML diagnosis, prognosis and development. Through high-throughput sequencing, hsa_circ_0006010 (circDPY19L1P1) was identified as a circRNA upregulating in AML. Thus, circDPY19L1P1 may exert a vital impact on AML development. Nevertheless, no relevant articles have revealed molecular mechanism of circDPY19L1P1 underlying AML.
Thus, herein, we attempted to elucidate circDPY19L1P1 role underlying AML. Our research may provide a novel insight for seeking targeted therapy plans of AML.
Research subjects
A total of 22 bone marrow specimens were obtained from AML patients, and 10 bone marrow specimens from Idiopathic Thrombocytopenic Purpura (ITP) patients were taken as negative controls. All participants in our research signed informed consent, and the ethics committee of our hospital granted approval for our research.
Cell lines and cell culture
Human AML cell lines (Kasumi-6 and Kasumi-3) and 293T cell line provided by ATCC (USA) received culture in Dulbecco's modified Eagle medium (DMEM; Hyclone, USA), 100 U/ml penicillin and 100 U/ml streptomycin at 37°C with 5% CO2, and cells at logarithmic growth phase were taken for further use.
RNA extraction and RT-qPCR
Total RNA received extraction from cells using TRIzol. mRNA and circRNA were reverse-transcribed through PrimeScript RT master mix and designed primers normalizing to GAPDH. Reverse transcriptions for miRNA were carried out through Bulge-Loop™ miRNA PCR starter kit and unique stem-loop primers normalizing to U6. The PCR reaction was run in triplicate through 7500 Real- Time PCR System with SYBR Premix Ex Taq II.
RNase R and actinomycin D treatment
For RNase R treatment, total RNA (2 μg) received incubation with or without RNase R (3 U/μg) at 37°C for 1 h. For blocking transcription, actinomycin D (2 mg/ml) or DMSO was added into cell culture medium. After treatment with actinomycin D and RNase R, RT-qPCR determined mRNA and circRNA expression levels in Kasumi-6 and Kasumi-3 cells.
Fluorescence in situ hybridization
The FITC-labeled circDPY19L1P1 probes were obtained from Geneseed Biotechnology (Guangzhou, China). Kasumi-6, Kasumi-3 and 293T cells were placed on coverslips, and received fixation, permeabilization in PBS with 0.5% Triton X-100 and dehydration in ethanol. The Fluorescence In Situ Hybridization (FISH) probes received dilution (1:50), denaturation and balancing, and were added to cells at 37°C overnight. Cells received labelling with DAPI-antifade for 10 min at room temperature post hybridization. Slides received sealing with rubber cement, placing in the dark for over 20 min, and detection through fluorescence microscopy (Leica, Switzerland).
Cell transfection
Short harpin RNA (shRNA) targeting circDPY19L1P1 (shcircDPY19L1P1# 1/2) or shRNA Negative Control (sh-NC), miR- 130a-3p mimics and Negative Control (NC mimics), PNPLA6 overexpression vector (pcDNA-PNPLA6) were obtained from GenePharma (Shanghai, China). Kasumi-6 and Kasumi-3 cells received culture to approximately 80% confluence in plates and then received transfection with designated plasmids using Lipofectamine 3000 under manufacturer's instructions. After 48 h, cells were harvested for further use.
EdU staining
EdU cell proliferation assay kit (Ribobio) was applied for assessing cell proliferation. The 100 μL of EdU (50 μM) was added per well, and Kasumi-6 and Kasumi-3 cells received culture for 2 h. Cell nuclei received staining with Hoechst (1 µg/mL) for 30 min. The cell proportion incorporated EdU received confirmation through fluorescence microscopy.
TUNEL staining
After indicated treatment, Kasumi-6 and Kasumi-3 cells received fixation using 4% formaldehyde for 15 min and staining using a TUNEL Bright-Red Apoptosis Detection kit (Vazyme, USA). TUNEL-positive cells received observation through fluorescence microscopy.
Western blotting
Total protein got extraction from AML cells using RIPA buffer containing protease and phosphatase inhibitors. Protein got separation by SDS-PAGE and then were transferred to PVDF membranes. Primary antibodies used in our research including anti-CD11b, anti-CD14, anti-PNPLA6 and anti-GAPDH as well as horseradish peroxidase (HRP)-conjugated secondary antibodies were provided by Abcam (Shanghai, China). Protein bands got visualization and detection through enhanced chemiluminescence.
RNA pulls down
The mixture of protein from Kasumi-6 and Kasumi-3 cells and 50 pmol of biotinylated miR-130a-3p (Bio-miR-130a-3p) or biotinylated arbitrary single nucleotide (Bio-NC) received incubation for 1 h with 50 µL of streptavidin agarose beads (Life Technologies) at 4°C. The associated RNA-protein complexes received elution with biotin elution buffer and boiling in SDS for 10 min. The elute received examination with RT-qPCR.
Statistical analysis
Statistical analysis was conducted using SPSS 22.0. Comparison between groups was performed using t-test. Comparison of three or more groups was carried out via one-way analysis of variance. Pearson correlation assessed association of circDPY19L1P1, miR- 130a-3p and PNPLA6 levels in AML bone marrow specimens. All assays were carried out in triplicate. The difference was statistically significant once P<0.05.
Upregulation of circDPY19L1P1 in AML and verification of the circular characteristics of circDPY19L1P1
Through high-throughput sequencing, circDPY19L1P1 was identified as a circRNA upregulating in AML. While its specific role underlying AML remains elusive. First, RT-qPCR measured circDPY19L1P1 expression status in AML bone marrow specimens and normal controls. As a result, circDPY19L1P1 presented a remarkable upregulation in bone marrow specimens relative to controls (Figure 1A). Additionally, through CircInteractome, we discovered that circDPY19L1P1 originated from back splicing by exon 8 and 9 of DPY19L1P1 pre-mRNA (Figure 1B). As we know, circRNAs are resistant to RNase R stimulation, thus we conducted RNase R digestion assay in AML cells. Linear RNA transcript (DPY19L1P1 mRNA) exhibited a marked degradation relative to circDPY19L1P1 in Kasumi-6 and Kasumi-3 cells, further confirming circular nature of circDPY19L1P1 transcript (Figure 1C). Furthermore, stability of two transcripts were evaluated under administration with transcription inhibitor, actinomycin D. RTqPCR depicted that half-life of DPY19L1P1 mRNA was 48 h while that of circDPY19L1P1 exceeded 48 h in Kasumi-6 and Kasumi-3 cells Figure 1D demonstrating that circDPY19L1P1 transcript presented more stability than linear transcript. RT-qPCR measured circDPY19L1P1 level in human AML cell lines (Kasumi-6 and Kasumi-3) and control cell line (293T). As a result, circDPY19L1P1 presented a remarkable upregulation in AML cells relative to controls (Figure 1E). Subsequently, we assessed localization of circDPY19L1P1 in AML cells. Through FISH, circDPY19L1P1 presented predominately cytoplasmic distribution in Kasumi-6 and Kasumi-3 cells whereas showed no marked distribution in 293T cells (Figure 1F). Collectively, circDPY19L1P1 presents upregulation and stable circular characteristics in AML cells.
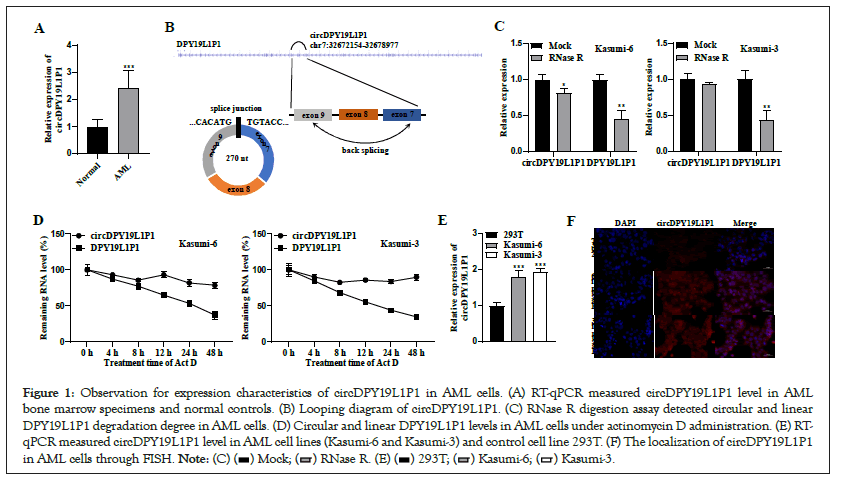
Figure 1: Observation for expression characteristics of circDPY19L1P1 in AML cells. (A) RT-qPCR measured circDPY19L1P1 level in AML bone marrow specimens and normal controls. (B) Looping diagram of circDPY19L1P1. (C) RNase R digestion assay detected circular and linear DPY19L1P1 degradation degree in AML cells. (D) Circular and linear DPY19L1P1 levels in AML cells under actinomycin D administration. (E) RT-qPCR measured circDPY19L1P1 level in AML cell lines (Kasumi-6 and Kasumi-3) and control cell line 293T. (F) The localization of circDPY19L1P1 in AML cells through FISH. 
CircDPY19L1P1 silencing suppresses proliferation, promotes apoptosis and blocks differentiation of AML cells
Due to circDPY19L1P1 upregulation in AML cells, we hypothesized that circDPY19L1P1 may function as an oncogene in AML cellular phenotypes. Thus, we carried out loss-of-function assays. First, shRNA vectors (sh-circDPY19L1P1#1/2) and control sh-NC were transfected into Kasumi-6 and Kasumi-3 cells. CircDPY19L1P1 level in AML cells presented a successful silencing after sh-circDPY19L1P1#1/2 transfection (Figure 2A). Then, EdU staining assessed AML cell proliferative ability. As a result, circDPY19L1P1 depletion resulted in a remarkable EdU-positive cell proportion reduction in Kasumi-6 and Kasumi-3 cells Figure 2B, suggesting that circDPY19L1P1 knockdown suppresses AML cell proliferation. Moreover, TUNEL staining assessed AML cell apoptosis. As a result, circDPY19L1P1 silencing facilitated TUNELpositive cell proportion elevation in Kasumi-6 and Kasumi-3 cells Figure 2C, suggesting that circDPY19L1P1 deficiency facilitates AML cell apoptosis. Additionally, AML is characterized by blocked differentiation of immature myeloid cells. CD11b and CD14 are classic differentiation-associated markers in leukemia [13]. Thus, we assessed cell CD11b and CD14 protein levels through western blotting. The results illustrated that circDPY19L1P1 knockdown caused CD11b and CD14 upregulation in Kasumi-6 and Kasumi-3 cells Figure 2D, suggesting that circDPY19L1P1 downregulation blocks AML cell differentiation. Collectively, circDPY19L1P1 acts as an oncogene through facilitating AML cell malignant behaviors.
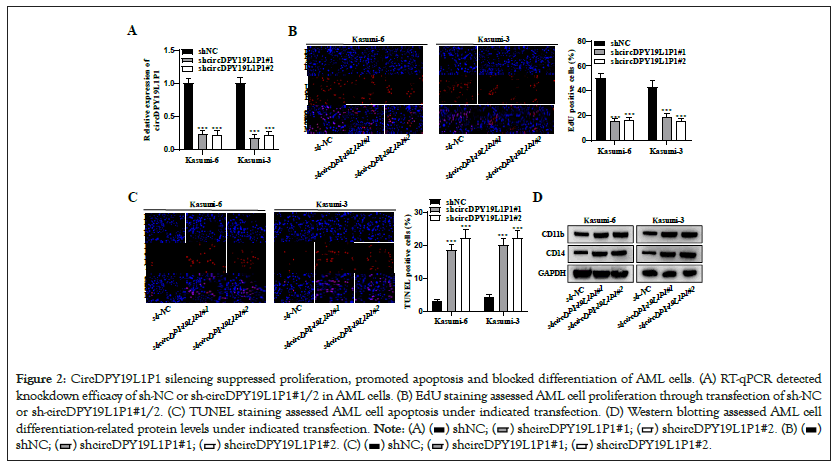
Figure 2: CircDPY19L1P1 silencing suppressed proliferation, promoted apoptosis and blocked differentiation of AML cells. (A) RT-qPCR detected knockdown efficacy of sh-NC or sh-circDPY19L1P1#1/2 in AML cells. (B) EdU staining assessed AML cell proliferation through transfection of sh-NC or sh-circDPY19L1P1#1/2. (C) TUNEL staining assessed AML cell apoptosis under indicated transfection. (D) Western blotting assessed AML cell differentiation-related protein levels under indicated transfection. 
CircDPY19L1P1 downregulates miR-130a-3p expression
CircRNAs usually "sponge" specific miRNAs via their binding sequences with miRNAs and competitively degrade inhibitory impact of miRNAs on their downstream targets, thereby modulating target gene level [14]. We hypothesized that circDPY19L1P1 may exert its role underlying AML cells in such manner. Through starBase, we obtained miR-130a- 3p which ranked the first in prediction results as downstream miRNA of circDPY19L1P1. RT-qPCR depicted that miR-130a-3p presented downregulation in AML bone marrow specimens relative to normal controls (Figure 3A). Pearson correlation analysis illustrated that circDPY19L1P1 level possessed a negative association with miR-130a-3p level in AML bone marrow specimens (Figure 3B). RT-qPCR measured miR-130a-3p level in human AML cell lines (Kasumi-6 and Kasumi-3) and control cell line (293T). As a result, miR-130a-3p presented a remarkable downregulation in AML cells relative to controls (Figure 3C). Through bioinformatics, we obtained binding sequence of miR-130a-3p on circDPY19L1P1 (Figure 3D). Simultaneously, miR-130a-3p level showed upregulation under sh-circDPY19L1P1#1/2 transfection in Kasumi-6 and Kasumi-3 cells Figure 3E, suggesting that circDPY19L1P1 exerts negative modulation on miR-130a-3p in AML cells. RNA pull down depicted that circDPY19L1P1 markedly enriched in complexes pulled-down by Bio-miR-130a-3p in Kasumi-6 and Kasumi-3 cells Figure 3F, supporting a binding of circDPY19L1P1 to miR-130a-3p in AML cells. Collectively, circDPY19L1P1 serves as a miR-130a-3p sponge to downregulate miR- 130a-3p in AML cells.
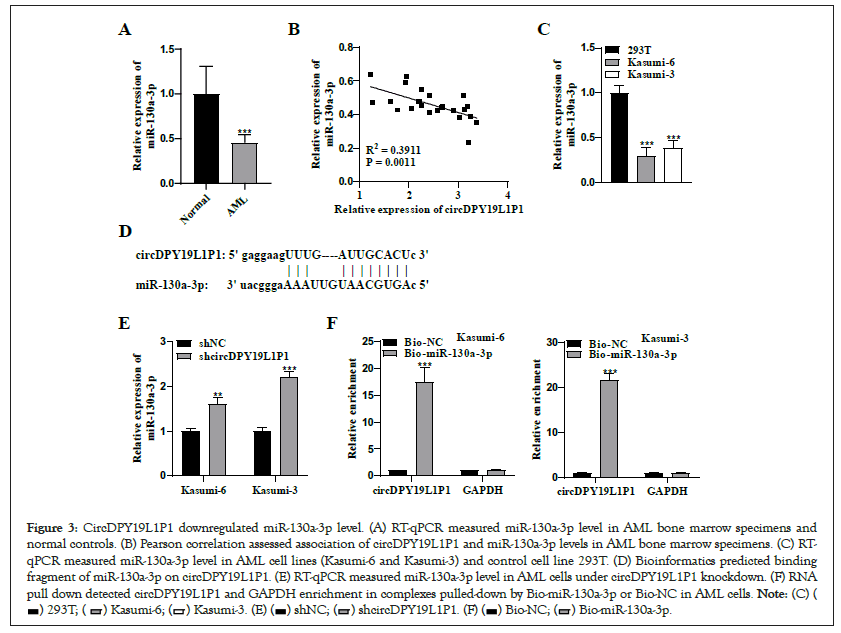
Figure 3: CircDPY19L1P1 downregulated miR-130a-3p level. (A) RT-qPCR measured miR-130a-3p level in AML bone marrow specimens and normal controls. (B) Pearson correlation assessed association of circDPY19L1P1 and miR-130a-3p levels in AML bone marrow specimens. (C) RT-qPCR measured miR-130a-3p level in AML cell lines (Kasumi-6 and Kasumi-3) and control cell line 293T. (D) Bioinformatics predicted binding fragment of miR-130a-3p on circDPY19L1P1. (E) RT-qPCR measured miR-130a-3p level in AML cells under circDPY19L1P1 knockdown. (F) RNA pull down detected circDPY19L1P1 and GAPDH enrichment in complexes pulled-down by Bio-miR-130a-3p or Bio-NC in AML cells. 
CircDPY19L1P1 positively regulates miR-130a-3p expression by PNPLA6
CeRNAs are transcripts post-transcriptionally modulating each other via competing for common miRNAs [15]. To further validate whether circDPY19L1P1 exerted a function via ceRNA pattern, starBase predicted candidate targets of miR-130a-3p with certain conditions. PNPLA6 ranked the first in prediction results. GEPIA depicted that PNPLA6 presented upregulation in AML specimens relative to controls (Figure 4A). Additionally, high-level PNPLA6 had a close relation to longer survival time of AML patients (Figure 4B). RT-qPCR depicted that PNPLA6 presented upregulation in AML bone marrow specimens relative to normal controls (Figure 4C). Pearson correlation analysis illustrated that PNPLA6 level possessed a negative association with miR-130a-3p level and a positive relation to circDPY19L1P1 level in AML bone marrow specimens (Figure 4D). RT-qPCR measured PNPLA6 level in human AML cell lines (Kasumi-6 and Kasumi-3) and control cell line (293T). As a result, PNPLA6 presented a remarkable upregulation in AML cells relative to controls (Figure 4E). Through bioinformatics, we obtained binding sequence of miR-130a-3p on PNPLA6 3'UTR (Figure 4F). RNA pull down depicted that PNPLA6 markedly enriched in complexes pulled-down by Bio-miR-130a-3p in Kasumi-6 and Kasumi-3 cells Figure 4G, supporting a binding of PNPLA6 to miR-130a-3p in AML cells. Through RT-qPCR and western blotting, we discovered that PNPLA6 mRNA and protein levels showed downregulation under sh-circDPY19L1P1#1/2 or miR-130a-3p mimics transfection in Kasumi-6 and Kasumi-3 cells Figure 4H, suggesting that circDPY19L1P1 exerts positive modulation on PNPLA6 whereas miR-130a-3p exerts negative modulation on PNPLA6 in AML cells. Collectively, miR-130a- 3p targets PNPLA6 3'UTR in AML cells and circDPY19L1P1 competitively binds to miR-130a-3p to upregulate PNPLA6.
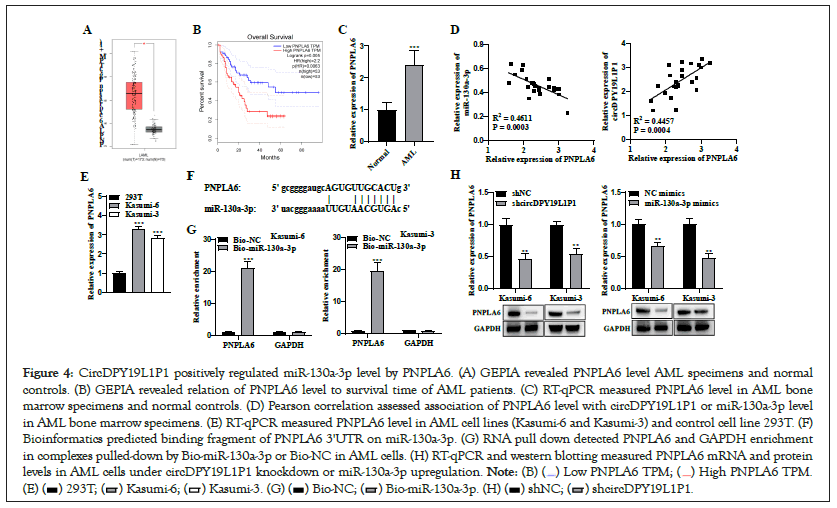
Figure 4: CircDPY19L1P1 positively regulated miR-130a-3p level by PNPLA6. (A) GEPIA revealed PNPLA6 level AML specimens and normal controls. (B) GEPIA revealed relation of PNPLA6 level to survival time of AML patients. (C) RT-qPCR measured PNPLA6 level in AML bone marrow specimens and normal controls. (D) Pearson correlation assessed association of PNPLA6 level with circDPY19L1P1 or miR-130a-3p level in AML bone marrow specimens. (E) RT-qPCR measured PNPLA6 level in AML cell lines (Kasumi-6 and Kasumi-3) and control cell line 293T. (F) Bioinformatics predicted binding fragment of PNPLA6 3'UTR on miR-130a-3p. (G) RNA pull down detected PNPLA6 and GAPDH enrichment in complexes pulled-down by Bio-miR-130a-3p or Bio-NC in AML cells. (H) RT-qPCR and western blotting measured PNPLA6 mRNA and protein levels in AML cells under circDPY19L1P1 knockdown or miR-130a-3p upregulation. 
PNPLA6 is essential for circDPY19L1P1 functioning in proliferation, apoptosis, and differentiation of AML cells
To elucidate PNPLA6 role in circDPY19L1P1-mediated AML cellular phenotypes, we conducted rescue experiments. PNPLA6 presented downregulation due to circDPY19L1P1 knockdown and presented upregulation after co-transfection with pcDNA-PNPLA6 vector (Figure 5A). We discovered that the depleted proliferative ability Figure 5B and elevated apoptosis Figure 5C as well as the depleted cell differentiation Figure 5D due to circDPY19L1P1 silencing were all rescued through PNPLA6 elevation. Collectively, circDPY19L1P1 facilitates AML cell malignant phenotypes via PNPLA6 upregulation.
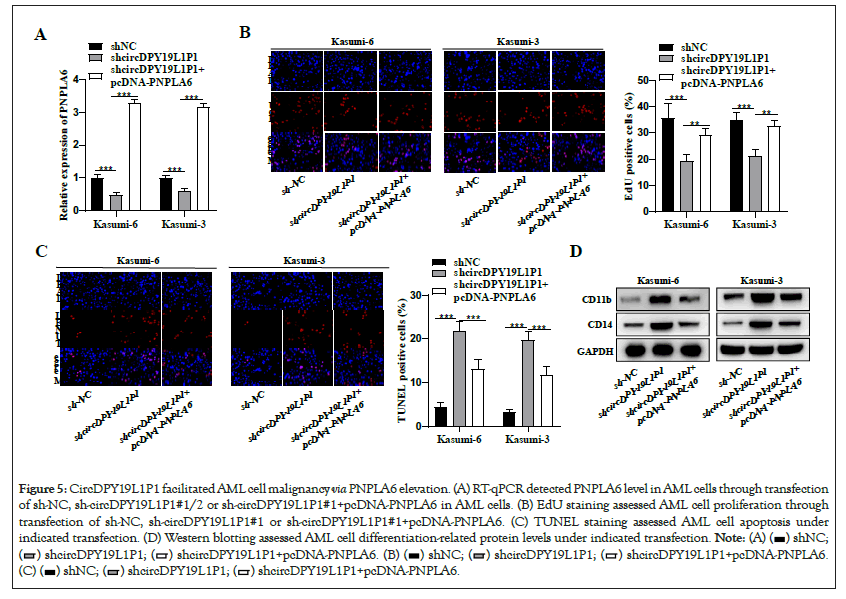
Figure 5: CircDPY19L1P1 facilitated AML cell malignancy via PNPLA6 elevation. (A) RT-qPCR detected PNPLA6 level in AML cells through transfection of sh-NC, sh-circDPY19L1P1#1/2 or sh-circDPY19L1P1#1+pcDNA-PNPLA6 in AML cells. (B) EdU staining assessed AML cell proliferation through transfection of sh-NC, sh-circDPY19L1P1#1 or sh-circDPY19L1P1#1+pcDNA-PNPLA6. (C) TUNEL staining assessed AML cell apoptosis under indicated transfection. (D) Western blotting assessed AML cell differentiation-related protein levels under indicated transfection. 
AML is a hematological malignancy characterized by abnormal proliferation of immature myeloid cells [16]. Clarifying molecular mechanism underlying AML pathogenesis and related genes and figuring out potential and effective biomarkers of AML is particularly crucial for improving prognosis of AML.
CircRNAs present wide expression in hematopoietic cells, and circRNA expression status can change with differentiation, with cell type specificity, which are regulators of hematopoietic cell differentiation, maturation and function [17,18]. CircRNAs not only get involvement in occurrence, development and prognosis of AML and other hematologic malignancies, but also have a more diverse expression profile in hematologic tumor cells relative to in normal cells, which affects protein expression in hematopoietic cells at different stages of development [19,20]. Through highthroughput sequencing, circDPY19L1P1 was identified as a circRNA upregulating in AML. Herein, circDPY19L1P1 presented a remarkable upregulation in bone marrow specimens relative to controls; circDPY19L1P1 presented a remarkable upregulation in AML cells relative to controls. Moreover, circDPY19L1P1 transcript presented more stability than linear transcript and presented predominately cytoplasmic distribution in AML cells. These findings suggested that circDPY19L1P1 presents upregulation and stable circular characteristics in AML cells. Additionally, circDPY19L1P1 knockdown suppressed AML cell proliferation facilitated AML cell apoptosis and blocked AML cell differentiation. These findings suggested that circDPY19L1P1 acts as an oncogene through facilitating AML cell malignant behaviors.
CircRNAs can indirectly modulate gene level through blocking binding of miRNAs to specific gene 3'UTR and exert a "spongelike" role [21]. Multiple reports on hematological malignancies have demonstrated that circRNAs may modulate occurrence, development and drug resistance of tumor cells through sponging miRNAs [22-24]. Thus, we hypothesized that circDPY19L1P1 may exert its role underlying AML cells in such manner. Herein, we obtained miR-130a-3p as downstream miRNA of circDPY19L1P1. Previously, miR-130a-3p suppresses bladder cancer lymphangiogenesis through VEGF-D pathway [25]. MiR-130a-3p hinders colorectal cancer progression through directly targeting WNT1 [26]. These reports indicate that miR-130a-3p may exert an inhibitory impact on tumors. Herein, miR-130a-3p presented a remarkable downregulation in AML bone marrow specimens relative to normal controls; miR-130a-3p presented a remarkable downregulation in AML cells relative to controls. Moreover, miR- 130a-3p level had a negative association with circDPY19L1P1 level in AML bone marrow specimens and circDPY19L1P1 silencing resulted in miR-130a-3p downregulation in AML cells. These findings suggested that circDPY19L1P1 exerts negative modulation on miR-130a-3p level. Furthermore, circDPY19L1P1 possessed binding sequence with miR-130a-3p and mechanism assays illustrated that circDPY19L1P1 bound to miR-130a-3p in AML cells. These findings suggested that circDPY19L1P1 serves as a miR-130a-3p sponge to downregulate miR-130a-3p in AML cells.
PNPLA6 belongs to a protein family containing nine patatin-like phospholipesterase domains [27]. With advancement of gene detection methods, the protein encoded by PNPLA6 gene has been proved to be a vital protein related to multiple diseases [28-30]. Nevertheless, PNPLA6 role in tumors including AML have never been clarified. Herein, we suspected that circDPY19L1P1 exerted a function via ceRNA pattern, and PNPLA6 was identified as putative target of miR-130a-3p. We discovered that PNPLA6 presented a remarkable upregulation in AML bone marrow specimens relative to normal controls; PNPLA6 presented a remarkable upregulation in AML cells relative to controls.
Moreover, PNPLA6 level had a positive association with circDPY19L1P1 level and possessed a negative relation to miR-130a- 3p level in AML bone marrow specimens; circDPY19L1P1 silencing or miR-130a-3p upregulation resulted in PNPLA6 downregulation in AML cells. These findings suggested that circDPY19L1P1 exerts positive modulation whereas miR-130a-3p exerts negative modulation on PNPLA6 level. Furthermore, PNPLA6 3'UTR possessed binding sequence with miR-130a-3p and mechanism assays illustrated that PNPLA6 bound to miR-130a-3p in AML cells. These findings suggested that miR-130a-3p targets PNPLA6 3'UTR in AML cells and circDPY19L1P1 competitively binds to miR-130a-3p to upregulate PNPLA6. Additionally, rescue assays demonstrated that circDPY19L1P1 facilitated AML cell malignant phenotypes via PNPLA6 upregulation.
In conclusion, circDPY19L1P1 presents upregulation and acts as an oncogene in AML cellular behaviors. CircDPY19L1P1 facilitates AML cell malignancy through serving as miR-513a-5p sponge to upregulate PNPLA6, providing a potential new direction for therapeutic strategies of AML. Nevertheless, animal assays should be performed in future research, further validating current conclusions.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Xu N, Ma R, Zhai H, Wang H (2023) CircDPY19L1P1/miR-130a-3p/PNPLA6 Axis: Regulating the Cell Differentiation in Acute Myeloid Leukemia (AML). Chemo Open Access. 11:172.
Received: 24-Feb-2023, Manuscript No. CMT-23-21924; Editor assigned: 01-Mar-2023, Pre QC No. CMT-23-21924 (PQ); Reviewed: 17-Mar-2023, QC No. CMT-23-21924; Revised: 24-Mar-2023, Manuscript No. CMT-23-21924 (R); Published: 31-Mar-2023 , DOI: 10.35248/2167-7700.23.11.172
Copyright: © 2023 Xu N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.