Journal of Infectious Diseases & Preventive Medicine
Open Access
ISSN: 2329-8731
ISSN: 2329-8731
Short Communication - (2022)Volume 10, Issue 2
The replication and transcription of the positive-sense single-stranded RNA genomes of SARS-CoV-1 and 2 require both continuous genomic synthesis and discontinuous subgenomic synthesis of messenger RNAs as is the case for all nidoviruses. To create the nested set of messenger RNAs, the elongating negative-sense strand jumps from a set of transcription regulatory sequences (TRS-Bs) to a similar leader transcription regulatory sequence (TRS-L) located in the 5’ genomic terminus. This unique mode of messenger RNA synthesis has prompted the speculation for the need for the proximity of the 5’ and 3’ termini during the synthesis of subgenomic negative-sense strands. Here, we report the presence of complementary terminal segments in SARS-CoV-1 and 2 that could mediate such circularization.
Transcription; Coronaviruses; Replication; Genomes; Subgenomic
Replication and transcription of coronaviruses is complex, requiring both continuous and discontinuous negative-strand synthesis in Figure 1 [1]. Circularization of the coronavirus genome which places the 5’ with 3’ termini and their regulatory sequences [2,3] in proximity has been postulated as a necessary early requirement for synthesis of subgenomic negative-sense strands [4]. We searched for and found the complementary sequences in the genomic termini of SARS-CoV-1 and 2 that could mediate genome circularization.
Nucleotide sequences were obtained from GenBank® and the Global Initiative on Sharing All Influenza Data (GISAID). Complementary sequences between genomic termini were visualized using forna,, a force directed graph layout (ViennaRNA Web services) [5]. Nucleotide sequences identified using the reference strains for SARS-CoV-2 (Wuhan reference strain, Genbank accession No. NC_045512) and SARS-CoV-1 (NC_004718) were searched and aligned with those of SARSCoV- 1 and 2 variants and other human beta coronaviruses using the Basic Alignment Search Tool (BLAST® , National Center for Biotechnology Information).
To estimate the free energy of the double-stranded structure formed by the complementary sequences, the sequences identified in the 5’ and 3’ genomic termini were analyzed as a single nucleotide sequence as if they were contiguous. Two stochastic folding simulations were used, one in the Sfold server (http://sfold.wadsworth.org and http://www.bioinfo.rpi.edu/ applications/sfold) [6] which is based on RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble [7]; and the other in the Kinefold server (http:// kinefold.curie.fr) for RNA/DNA folding path and structure prediction including pseudoknots and knots [8-14].
The present analysis revealed two complementary segments in the genomic termini of SARS-CoV-1 and SARS-CoV-2 (Figure 1). The 5’ sequence is 36 nucleotides long and begins at position 60 from the 5’ terminal m7G cap structure of SARS-CoV-2. This 36-nucleotide segment includes the stem-loop structure SL3, which encompasses the leader transcription regulatory sequence (TRS-L: ACGAAC) in the 5’ leader sequence. We designate this as the 5’ complementary circularization sequence (5’CCS). The 5’CCS extends to the beginning of stem-loop 4 (SL4) (Figure 2). A 36-nucleotide sequence complementary to the 5’CCS is located at the 3’ terminus immediately 5’ to the site of polyadenylation (Figure 3). We designate this as the 3’ complementary circularization sequence (3’CCS). The 5’CSS and 3’CCS are conserved in all SARS-CoV-1 and 2 sequences deposited in GenBank and GISAID with one exception. The guanosine at position 90 of the 5’CCS of SARS-CoV-2 is replaced by adenosine at the corresponding position of SARSCoV- 1 isolates (underlined in Figure 1). The CCSs are not present in other human beta coronaviruses we have examined.
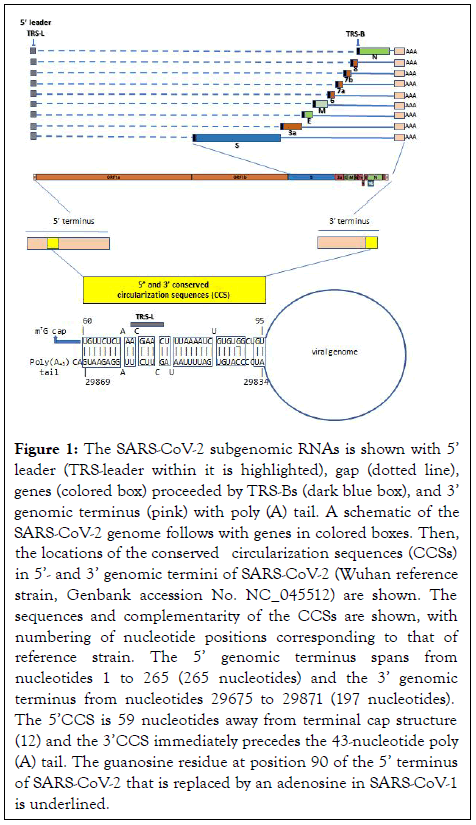
Figure 1: The SARS-CoV-2 subgenomic RNAs is shown with 5’ leader (TRS-leader within it is highlighted), gap (dotted line), genes (colored box) proceeded by TRS-Bs (dark blue box), and 3’ genomic terminus (pink) with poly (A) tail. A schematic of the SARS-CoV-2 genome follows with genes in colored boxes. Then, the locations of the conserved circularization sequences (CCSs) in 5’- and 3’ genomic termini of SARS-CoV-2 (Wuhan reference strain, Genbank accession No. NC_045512) are shown. The sequences and complementarity of the CCSs are shown, with numbering of nucleotide positions corresponding to that of reference strain. The 5’ genomic terminus spans from nucleotides 1 to 265 (265 nucleotides) and the 3’ genomic terminus from nucleotides 29675 to 29871 (197 nucleotides). The 5’CCS is 59 nucleotides away from terminal cap structure (12) and the 3’CCS immediately precedes the 43-nucleotide poly (A) tail. The guanosine residue at position 90 of the 5’ terminus of SARS-CoV-2 that is replaced by an adenosine in SARS-CoV-1 is underlined.
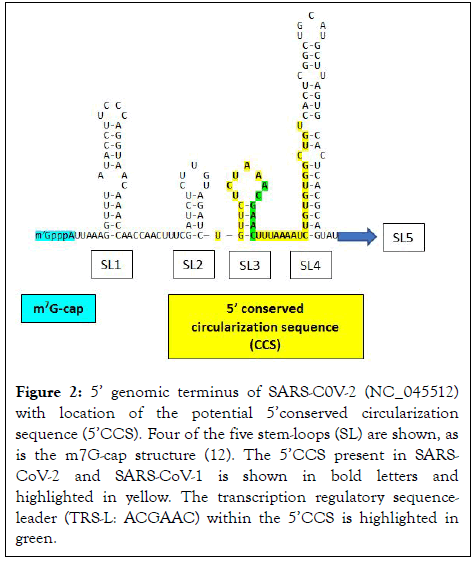
Figure 2: 5’ genomic terminus of SARS-C0V-2 (NC_045512) with location of the potential 5’conserved circularization sequence (5’CCS). Four of the five stem-loops (SL) are shown, as is the m7G-cap structure (12). The 5’CCS present in SARSCoV- 2 and SARS-CoV-1 is shown in bold letters and highlighted in yellow. The transcription regulatory sequenceleader (TRS-L: ACGAAC) within the 5’CCS is highlighted in green.
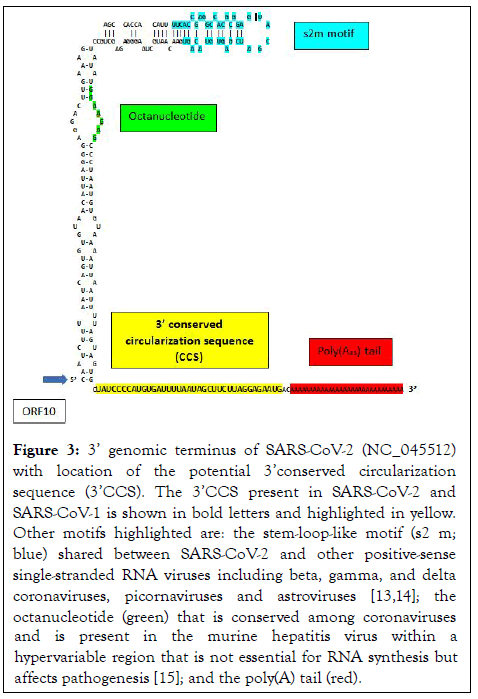
Figure 3: 3’ genomic terminus of SARS-CoV-2 (NC_045512) with location of the potential 3’conserved circularization sequence (3’CCS). The 3’CCS present in SARS-CoV-2 and SARS-CoV-1 is shown in bold letters and highlighted in yellow. Other motifs highlighted are: the stem-loop-like motif (s2 m; blue) shared between SARS-CoV-2 and other positive-sense single-stranded RNA viruses including beta, gamma, and delta coronaviruses, picornaviruses and astroviruses [13,14]; the octanucleotide (green) that is conserved among coronaviruses and is present in the murine hepatitis virus within a hypervariable region that is not essential for RNA synthesis but affects pathogenesis [15]; and the poly(A) tail (red).
The free energy of the double-strand formed by the complementary genomic termini sequences of SARS-CoV-1 and 2 ranged from -18.70 Kcal/mol at 37°C for the centroid structure generated by Boltzmann-weighted structure (Sfold server) to -15.8 Kcal/mol as per the Kinefold server simulation.
The present finding of complementary nucleotide sequences in the 5’ and 3’ termini of the genomes of SARS-CoV-1 and 2 is consistent with the hypothesis of circularization as a necessary early step in replication and subgenomic RNA synthesis and points to possible involvement of a direct RNA-RNA interaction.
SARS-CoV-1 caused an epidemic in 2002 with 8437 cases in 30 countries, and SARS-CoV-2 a pandemic since 2019 and approaching 300 million cases globally. The finding of potential circularization sequences only in the termini of SARS-CoV-1 and 2 RNA genomes raises the question of how other human coronaviruses replicate without 5’ and 3’CCSs sequences. A protein bridge composed of cap binding protein, eIF4E, eIF4G, and poly (A) binding protein may mediate circularization and subsequent initiation of replicase gene translation [9-15]. We speculate that the presence of the circularization sequences in SARS-CoV-1 and 2 may help stabilize such protein bridges and potentially translate into increased replication competence of SARS-CoV-1 and 2.
In bovine coronavirus, the N protein can act as a bridge to facilitate interaction between the 5'- and 3'-ends of its genome, leading to circularization of the genome [11]. It remains to be determined which viral and cellular factors contribute to the replication complexes and how the putative RNA-RNA interaction described here contributes to facilitating or stabilizing RNA-protein and protein-protein interactions in subgenomic RNA synthesis.
Citation: Patarca R, Haseltine WA (2022) Circularization via Complementary Sequences in the 5â?? and 3â?? Termini may Facilitate Replication of SARS Coronaviruses. J Infect Dis Preve Med. 10: 258.
Received: 14-Feb-2022, Manuscript No. JADPR-22-15852; Editor assigned: 17-Feb-2022, Pre QC No. JADPR-22-15852 (PQ); Reviewed: 04-Mar-2022, QC No. JADPR-22-15852; Revised: 10-Mar-2022, Manuscript No. JADPR-22-15852 (R); Published: 17-Mar-2022 , DOI: 10.35841/2329-8731.22.10.258
Copyright: © 2022 Patarca R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.