Journal of Leukemia
Open Access
ISSN: 2329-6917
ISSN: 2329-6917
Research Article - (2017) Volume 5, Issue 4
This study enrolled 49 patients with de novo childhood acute lymphoblastic leukemia (ALL) and 19 adult patients with ALL. WT1 mRNA showed high positive expression rates of 90% or more in ALL patients, and fusion gene transcripts were detected in 22.4% of childhood ALL cases prior to treatment. During follow-up at 4-6 months, WT1 mRNA levels in childhood ALL cases were observed to be lower in patients undergoing hematological remission after treatment when compared to the initial stages. We also assessed hematological remission via ROC analysis. The upper cut-off values were calculated to be 220 and 1,820 copies/μg RNA in the peripheral blood (PB) and bone marrow (BM) samples, respectively. Since 50 copies/μg RNA was determined to be both, the limit of detection (LOD) and minimal residual disease (MRD) threshold, WT1 mRNA regions below the upper cut-off values indicated remission depth. The detection of high WT1 mRNA levels, even in patients without fusion gene transcripts, reflected the treatment effects and remission depth, demonstrating its capacity to be a useful MRD monitoring marker in ALL.
Keywords: WT1 mRNA; Reverse transcription-polymerase chain reaction (RT-PCR); Acute lymphoblastic leukemia (ALL); Minimal residual disease (MRD)
Acute lymphoblastic leukemia (ALL), a hematologic malignancy characterized by tumorigenic changes in the juvenile lymphoid cell and the bone marrow infiltration, has an annual incidence of approximately 1.7 in 100,000 children and adult per year [1]. ALL accounts for 70% of leukemia/malignant lymphoma in children, and it is estimated to be newly diagnosed in 550–600 children annually in Japan [2].
When selecting a treatment strategy to induce the remission of ALL/ Lymphoblastic Lymphoma (LBL), the presence of the Philadelphia (Ph) chromosome is initially used for classification purposes. While a positive Ph chromosome results in priority treatments with tyrosine kinase inhibitors such as imatinib, a pediatric protocol (Programa Español de Tratamiento en Hematología ALL-96) [3] has been recommended for adolescent or young adult patients (below 30 years of age) presenting as Ph chromosome-negative. In Europe and America, more than 2,000 patients included in the Berlin-Frankfurt-Münster, Medical Research Council or Children’s Oncology Group clinical studies have shown improved treatment outcomes via appropriate stratification [4].
Currently, treatment optimization has been attempted by quantifying and using minimal residual disease (MRD) for stratification. In the Japan Pediatric Leukemia Lymphoma Study Group (JPLSG), which was started in 2012, MRD has been used for stratification under the ALL-B12 protocol throughout Japan.
Monitoring MRD is important in predicting a patient’s disease relapse and optimizing the treatment strategy. While morphological examination can reveal a decrease in lymphoblasts in the bone marrow (BM) or peripheral blood (PB), indicating hematological remission, MRD cannot be detected by this method and therefore, molecular methods are employed. MRD in ALL patients has been measured and assessed using several techniques such as quantification of fusion gene transcripts [5-8], immunoreceptor (immunoglobulin/T-cell receptor (Ig/TCR)) gene rearrangements [9-12], and flow cytometry [8,13-15]. However, fusion gene transcripts are expressed in approximately 25% of adult ALL patients and 3% of childhood ALL patients, even in minor BCR/ABL, which is thought to have the highest expression frequency [16]. Even when other fusion gene transcripts are combined, fusion gene transcripts as an MRD monitoring marker can only be used in a small number of patients. MRD quantification via flow cytometry [8,17] requires professional skills and experience and is only used in medical (research) institutions. MRD quantification via Ig/TCR gene rearrangements requires a primer setting specific to the clone. Although the sensitivity of the technique is high, the associated cost is also high, and the proliferation needs to occur in the same clone. Recently, a nextgeneration sequencer identified cancerous cells to have multiple clones rather than just one. Based on this theory, relapse is thought to occur via the subclones tolerant to chemotherapy. This sequencer detects relapse with different gene rearrangements [18]. MRD detection and the use of bone marrow has also been investigated with this next-generation sequencer, but the associated cost was high [19].
Wilms’ tumor 1 (WT1) is expressed in high frequency in the PB and BM of many patients with acute myeloid leukemia (AML) [20,21]. The development of in vitro WT1 messenger RNA (mRNA) diagnostics has resulted in its wide use in clinical practice as an MRD monitoring marker. Interestingly, WT1 mRNA is also expressed in ALL patients as noted in the following reports. In adult ALL patients, immature B-ALL cases expressed higher levels of WT1 mRNA than B-ALL cases [22], and T-ALL cases expressed higher levels of WT1 mRNA than B-ALL cases [23]. Furthermore, WT1 mRNA levels in a positive group of myeloid markers (CD13, CD33, CD15, and CD65) were higher than those in a negative group [22]. Patients with high expressions of WT1 mRNA exhibited shorter overall survival and disease-free survival periods, and the increase in the WT1 mRNA level in the BM related to the significantly increased risk of relapse. Therefore, WT1 mRNA has been reported to be a useful indicator in predicting prognosis [23-25].
Based on the above findings, we investigated the clinical significance of WT1 mRNA as an MRD monitoring marker in ALL patients by measuring the PB and BM WT1 mRNA levels in childhood and adult ALL patients.
Patients and samples
Between December 2014 and December 2015, BM and PB samples from 49 children and 19 adult patients with de novo ALL were collected by the investigators from their respective medical institutions as shown in Table 1. PB and BM samples from each patient were collected on the same day and used for WT1 mRNA measurement. The experimental protocol was reviewed and approved by the appropriate local ethics committees.
| Classification for the immunophenotypic groups | No. of patients | |||
|---|---|---|---|---|
| Childhood | Adult | Total | ||
| B-lineage ALL | B-precursor ALL | 43 | 15 | 58 |
| pre-B ALL | 5 | 3 | 8 | |
| mature B ALL | 0 | 0 | 0 | |
| T-lineage ALL | 1 | 0 | 1 | |
| Unknown | 0 | 1 | 1 | |
| Total | 49* | 19 | 68 | |
ALL adult acute lymphoblastic leukemia
* In one out of 49 children, peripheral blood samples could not be collected before treatment.
Table 1: Immunophenotypic profile of 68 de novo cases of childhood and adult acute lymphoblastic leukemia by flow cytometric analysis in JPLSG.
Among the pediatric patients, 43 patients were diagnosed with B-precursor ALL, 5 with pre-B-ALL, and 1 with T-ALL according to the immunophenotypic analysis. Among the adult ALL patients, 15 were diagnosed with B-precursor ALL and 3 were diagnosed with pre- B-ALL. The subclass for 1 remaining adult ALL patients could not be determined due to the lack of the flow cytometry measurement.
Out of the 49 childhood ALL patients who underwent induction therapy, 1 patient was transferred and hence, follow up was not possible. The remaining 48 childhood ALL patients achieved hematological remission after induction therapy.
All childhood ALL patients followed the JPLSG ALL-B12 protocol, which consisted of a prophase therapy (prednisolone across all risks), an induction therapy (prednisolone, daunorubicin, L-asparaginase, intrathecal injection of prednisolone + methotrexate + cytarabine), and an early consolidation therapy (cyclophosphamide, cytarabine, 6-mercaptopurine for intermediate risk, randomization of vincristine + L-asparaginase for high risk, intrathecal injection of prednisolone + methotrexate + cytarabine) followed by a consolidation therapy and a maintenance therapy. We classified the follow-up period as 4–6 months after the start of the treatment and collected the BM and PB samples from childhood ALL patients at the following time points: 1) before treatment, 2) after induction therapy, 3) after early consolidation therapy, and 4) later stage of the early consolidation therapy. BM and PB samples from adult ALL patients were collected prior to the treatment. Informed consent was obtained from all patients above 20 years of age, or from a legal representative for patients 16 years and above but less than 20 years, or from the guardians of patients under the age of 16 years.
WT1 mRNA level
We used the following calculation method elucidated by Kitamura et al. [26] to measure the expression of WT1 mRNA: total RNA was extracted from the patient PB or BM samples using the QIAamp RNA Blood Mini kit (Qiagen, Valencia, CA, USA), and WT1 mRNA was determined using the WT1 mRNA OneStep Assay Kit (Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan) based on a one-step reverse transcription quantitative polymerase chain reaction (RT-qPCR). The PCR primer and TaqMan probe from this kit assigned to exon 6–7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a comparative control [26,27]. The WT1 mRNA expression levels were calculated by multiplying the value obtained after dividing the measured WT1 mRNA with the measured value of GAPDH mRNA (number of WT1 mRNA copies per copy of GAPDH mRNA) with the mean GAPDH mRNA measurement value per 1 μg of RNA (2.7 × 107 copies/μg RNA) based on independent tests in healthy adults [20]. The unit for WT1 mRNA expression was prescribed as copies/μg RNA. The method to calculate WT1 mRNA expression is shown below:
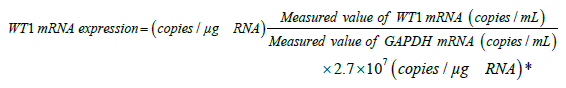
*2.7 × 107 (copies/μg-RNA): Mean GAPDH mRNA measurement value per 1 μg of RNA in a PB sample of a healthy adult.
Fusion gene transcripts
At the initial diagnosis, we screened for fusion gene transcripts. In this test, total RNA was extracted from the patient PB or BM samples and the following nine fusion gene transcripts were measured by RTqPCR: major BCR-ABL, minor BCR-ABL, E2A-PBX1, ETV6-AML1, MLL-AF4, MLL-AF6, MLL-AF9, MLL-ENL, and SIL-TAL1 [28]. Upon detection, we assayed the fusion gene transcripts and measured the WT1 mRNA levels [29,30].
Ig/TCR gene rearrangements
For the analysis of the rearranged immunoglobulin (Ig) and T-cell receptor (TCR) genes, qPCR was performed using standardized allelespecific oligonucleotide (ASO) primer/probe sets for the junctional regions of IgH, Igκ, TCRδ, TCRγ, TCRβ, or TAL1 [10]. Genomic DNA was isolated from the PB or BM samples using the QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA), and a 10-μL aliquot of the isolated DNA was subjected to qPCR involving 50 cycles with 0.25 μL of each ASO primer using a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Each PCR cycle included denaturation at 95°C for 15 s and primer annealing and extension at 60°C for 1 min, after pretreatment at 50°C for 2 min and 95°C for 10 min. MRD was identified by screening by multiplex PCR with allelespecific oligonucleotide primers. Then, the quantitative value of Ig/ TCR gene rearrangements was corrected using the albumin gene, which was simultaneously quantified as the internal control. The quantitative limit of the MRD targets in qPCR was 10−5 for the Ig/TCR gene rearrangements.
Flow cytometry
Ficoll-Hypaque-enriched blasts were stained by five-color immunofluorescence staining. The fluorophores phycoerythrin (PE), fluorescein isothiocyanate (FITC), phycoerythrin-cyanin 7 (PC7), or allophycocyanin (APC) were used in conjugation with the following antigens: CD1a, CD2, CD3, CD4, CD5, CD7, CD8, CD10, CD11b, CD13, CD14, CD15, CD19, CD20, CD21, CD22, CD24, CD33, CD34, CD36, CD38, CD41, CD42b, CD45, CD56, CD58, CD61, CD64, CD65, CD66c, CD99, CD117, Glycophorin-A (CD235a), HLA-DR, Immunoglobulin κ and λ light chains, T-cell receptor (TCR-α/β and TCR-γ/δ), cytoplasmic μ chain, cytoplasmic CD3, cytoplasmic CD22, cytoplasmic CD79a, cytoplasmic MPO, cytoplasmic TdT, and CRLF2. An isotype-matched immunoglobulin was used as the negative control. The analysis gate was set using CD45, for which low-blast gates were stained using various combinations of monoclonal antibodies. These antibody combinations used in this study were modified from those used in the study by Iwamoto et al. [31].
Classification of pediatric and adult ALL patients
The ALL patients were classified into immunophenotypic groups from the results obtained via flow cytometry according to the immunological diagnostic criteria of the JPLSG [31]. Definition of the four risk groups are as follows: B-precursor ALL (positive for 2 or more of CD19, CD20, CD22, or CD79a, and negative throughout the intracytoplasmic μ-chain, Igκ, and Igλ); pre-B ALL (positive for 2 or more of CD19, CD20, CD22, or CD79a, positive intracytoplasmic μ-chain, and negative Igκ and Igλ); mature B-ALL (positive for 2 or more of CD19, CD20, CD22, or CD79a, and positive Igκ or positive Igλ); and T-ALL (T-cell line).
Statistical analysis
For group comparison, logarithmic transformed values of WT1 mRNA levels (copies/μg RNA) were used for the Student’s t-test, Fisher’s exact test, two-way factorial analysis of variance, and Dunnett test or Tukey-Kramer honestly significant difference (HSD) test. Differences with p<0.05 were considered to be significant.
For the WT1 mRNA level, mean ± standard deviation was calculated with logarithmic transformed values of WT1 mRNA (copies/μg-RNA) and the mean was considered to be the value (back-transformed mean). Measured values below 50 copies/μg-RNA of WT1 mRNA were all treated as 49 copies/μg RNA. Additionally, the Pearson’s correlation coefficient was also calculated and evaluated.
Correlation between the PB and BM WT1 mRNA levels
Figure 1 shows the correlation between the PB and BM WT1 mRNA levels from all the measured samples. Samples from both, childhood ALL and adult ALL, demonstrated good correlation of WT1 mRNA values between BM and PB (correlation coefficient: r=0.85 and 0.84, respectively) and the regression lines were similar. The limit of detection (LOD) in PB was 50 copies/μg RNA, which was used as a threshold for MRD. Therefore, 260 copies/μg RNA of BM, which was obtained by substituting the LOD of PB into the correlation regression line equation, was considered to be the cut-off value to indicate BM abnormality. Subsequently, it was also specified as the cut-off value for WT1 mRNA positivity in the BM samples of ALL patients.
WT1 mRNA level and the positive rate in de novo ALL patients
Table 2 shows the WT1 mRNA levels and positive rates with respect to subtypes (B-precursor ALL and pre-B ALL) in the PB and BM samples of de novo ALL patients. The cut-off values for WT1 positive/ negative were determined as 50 copies/μg RNA for PB and 260 copies/ μg RNA for BM. In de novo childhood ALL patients, WT1 mRNA positivity was detected in the PB of 45 out of 48 patients (excluding one patient whose PB sample was not collected at diagnosis; 93.8%) and the BM of 39 out of 43 patients who were able to provide samples (90.7%). In de novo adult ALL patients, the positivity rate was 89.5% (17 out of 19 patients) for PB samples and 94.1% (16 out of 17 patients) for BM samples. The PB and BM sample from both, childhood and adult ALL patients, showed a high rate of positivity. With respect to the subtypes, PB samples from B-precursor ALL showed higher mean values than those from pre-B ALL in both, childhood and adult ALL patients (Figure 2a). The BM samples from B-precursor ALL also showed higher mean values than those from pre-B ALL (Figure 2b). Among the entire ALL subtype, the mean values of the PB samples from the B-precursor ALL was significantly higher than those from the pre-B ALL (p<0.05; Figure 2a).
| a. WT1 mRNA expression levels in PB | ||||||
| Group | B-precursor ALL | pre-B ALL | T ALL | unknown | Total | |
| Childhood ALL | No. of patients | 42 | 5 | 1 | - | 48 |
| Mean (range) (copies/µg RNA) | 1,100 (<50-140,000) | 270 (<50-3,300) | 91 | 910 (<50-140,000) | ||
| Positive rate (%) | 95.2 (40/42) | 80.0 (4/5) | 100 (1/1) | 93.8 (45/48) | ||
| Adult ALL | No. of patients | 15 | 3 | - | 1 | 19 |
| Mean (range) (copies/µg RNA) | 2,090 (<50-25,000) | 330 (<50-3,000) | 1,100 | 1,480 (<50-25,000) | ||
| Positive rate (%) | 93.3 (14/15) | 66.7 (2/3) | 100 (1/1) | 89.5 (17/19) | ||
| Total ALL | No. of patients | 57 | 8 | 1 | 1 | 67 |
| Mean (range) (copies/µg RNA) | 1,290 (<50-140,000) | 290 (<50-3,300) | 91 | 1,100 | 1,050 (<50-140,000) | |
| Positive rate (%) | 94.7 (54/57) | 75.0 (6/8) | 100 (1/1) | 100 (1/1) | 92.5 (62/67) | |
| b. WT1 mRNA expression in BM | ||||||
| Group | B-precursor ALL | pre-B ALL | T ALL | unknown | Total | |
| Childhood ALL | No. of patients | 38 | 4 | 1 | - | 43 |
| Mean (range) (copies/µg RNA) | 3,890 (90-180,000) | 800 (320-3,100) | 1,500 | 3,240 (90-180,000) | ||
| Positive rate (%) | 89.5 (34/38) | 100 (4/4) | 100 (1/1) | 90.7 (39/43) | ||
| Adult ALL | No. of patients | 14 | 3 | - | - | 17 |
| Mean (range) (copies/µg RNA) | 5,620 (650-45,000) | 1,260 (<50-14,000) | 4,270 (<50-45,000) | |||
| Positive rate (%) | 100 (14/14) | 66.7 (2/3) | 94.1 (16/17) | |||
| Total ALL | No. of patients | 52 | 7 | 1 | - | 60 |
| Mean (range) (copies/µg RNA) | 4,270 (90-180,000) | 960 (<50-14,000) | 1,500 | 3,310 (<50-180,000) | ||
| Positive rate (%) | 92.3 (54/57) | 85.7 (6/7) | 100 (1/1) | 91.7 (55/60) | ||
Numbers in parentheses indicate number of patients
Table 2: WT1 mRNA expression levels and WT1 mRNA-positive rate in PB and BM on initial untreated ALL patients.
Figure 2: Comparison among ALL immunophenotypes by Wilms’ tumor 1 (WT1) mRNA expression from childhood acute lymphoblastic leukemia (ALL) patients, adult ALL patients, and total ALL patients. a. peripheral blood; b. Bone marrow. Bold lines represent mean WT1 mRNA expression after log transformation. Fine line represents minimum limit of detection of WT1 mRNA (50 copies/μg RNA).
Changes in the WT1 mRNA levels during follow-up
Figure 3 shows the changes in the WT1 mRNA levels in the PB and BM samples from 48 childhood ALL patients during follow-up, except for the one transferred patient. All 48 patients were in the hematological remission phase at all blood collection points after treatment. WT1 mRNA levels were observed to decrease (in 40 out of 48 PB samples (83.3%) and in 35 out of 42 BM samples (83.3%); p < 0.01; Dunnett test) at all points of blood collection after treatment when compared to any point prior to treatment.
WT1 mRNA and fusion gene transcripts
Screening for fusion gene transcripts in childhood ALL patients was performed prior to treatment and 7 patients with ETV6-AML1, 2 patients with E2A-PBX1, 1 patient with minor BCR-ABL, and 1 patient with major BCR-ABL was detected. The detection rate was observed to be 22.4% (11 out of 49 samples), which was significantly lower than that of WT1 mRNA (93.8% in PB and 90.7% in BM, p<0.01; Fisher’s exact test). Figure 4 shows the changes in expression during follow-up in 11 patients with fusion gene transcripts. Moreover, the WT1 mRNA expression level in the BM and PB of all the patients tended to decrease to that of the fusion gene transcripts.
Figure 4: Transition between fusion gene transcript and Wilms’ tumor 1 (WT1) mRNA expression levels. a. minor BCR-ABL (n=1); b. major BCR-ABL (n=1; c. E2A/PBX1 (n=2); and d. ETV6-AML1 (n=7). Fine line represents minimum limit of detection of WT1 mRNA (50 copies/μg RNA). Changes in expression levels of fusion gene transcript in PB on the left side and BM on the right side were indicated by dotted lines for each detected type of fusion gene transcript, the transition of WT1 mRNA expression in the same patient was superimposed by solid line.
WT1 mRNA levels by hematological phases
Except for one patient who transferred during the follow-up, the other 48 childhood ALL patients were in the hematological remission phase at blood collection points after starting treatment. Therefore, we compared the WT1 mRNA levels at all blood collection points during the untreated and remission phases (Figure 5). As a result, WT1 mRNA levels during the remission phase were significantly lower than those during the untreated phase in both, PB and BM samples (p<0.01; Student’s t-test). To assess remission in terms of WT1 mRNA levels, we calculated the cut-off values of WT1 mRNA in both groups to differentiate the remission phase via receiver operating characteristic (ROC) analysis. Figure 6 shows the results of the analysis. The cutoff values obtained to determine remission were 220 copies/μg RNA for PB and 1,820 copies/μg RNA for BM. The -sensitivity and specificity for PB were determined to be 68.8% (33 out of 48 samples) and 98.0% (99 out of 101 samples) respectively, whereas the sensitivity and specificity for BM were determined to be 58.1% (25 out of 43 samples) and 98.0% (97 out of 99 samples) respectively.
WT1 mRNA and Ig/TCR gene rearrangements
The analysis of Ig/TCR gene rearrangements was performed using BM from five child patients with B-precursor ALL and one with Pre-B ALL at different time points before treatment, after induction, and after early consolidation. The gene rearrangements were detected in the BM of all six patients at the time point before treatment, and in four patients after induction. After early consolidation, the gene rearrangements were shown to be “positive, outside quantitative range” in two patients; however, the other four showed negative (Table 3).
| Patient No. | Analysis method for MRD | Sample | Time point for sample collection | ||
|---|---|---|---|---|---|
| Before treatment | After induction | After early consolidation |
|||
| No.02-001 (B-precursor ALL) |
WT1 mRNA | BM | 280 | 370 | 1,500 |
| PB | 60 | 130 | 180 | ||
| Ig/TCR gene Rearrangement |
BM | 1 | negative | negative | |
| No.02-004 (B-precursor ALL) |
WT1 mRNA | BM | 31,000 | 330 | 1,300 |
| PB | 21,000 | <50 | <50 | ||
| Ig/TCR gene rearrangement |
BM | 1 | 1.90×10-4 | positive, outside QR | |
| No.05-001 (B-precursor ALL) |
WT1 mRNA | BM | 76,000 | 230 | 490 |
| PB | 64,000 | 66 | <50 | ||
| Ig/TCR gene rearrangement |
BM | 1 | 9.20×10-5 | negative | |
| No.07-001 (Pre-B ALL) |
WT1 mRNA | BM | 640 | 130 | 270 |
| PB | 150 | 110 | 160 | ||
| Ig/TCR gene rearrangement |
BM | 1 | positive, outside QR | negative | |
| No.07-003 (B-precursor ALL) |
WT1 mRNA | BM | no sample | 290 | 1,500 |
| PB | 32,000 | 100 | <50 | ||
| Ig/TCR gene rearrangement |
BM | 1 | 1.60×10-4 | positive, outside QR | |
| No.07-004 (B-precursor ALL) |
WT1 mRNA | BM | 16,000 | 1,700 | 2,100 |
| PB | 5,800 | 190 | 100 | ||
| Ig/TCR gene rearrangement |
BM | 1 | negative | negative | |
Abbreviations: ALL, acute lymphoblastic leukemia; Ig, immunoglobulin; MRD, minimal residual disease; TCR, T-cell receptor; Positive, outside QR, Positive, outside quantitative range; BM, bone marrow; PB, peripheral blood. WT1 mRNA level was expressed as the copy number in a microgram of RNA; contrastingly, the quantitative value of Ig/TCR gene rearrangements was a value relative to the value when the patient visited the hospital at first, and was expressed as a power of 10 (×10-n). MRD targets with quantitative limits of <10-5 for qPCR for Ig/TCR gene rearrangements and <50 copies/μg RNA for RT-qPCR for WT1 mRNA.
Table 3: Correlation between WT1 mRNA expression levels and Ig/TCR gene rearrangements.
WT1 mRNA levels in the BM of all six patients after induction were determined to be lower than 1,820 copies/μg RNA, which was determined as the cut-off value in BM for remission, as described above. WT1 mRNA levels in the PB of all six patients at all time points were determined to be lower than 220 copies/μg RNA, which was determined as the cut-off value in PB for remission, as described above.
ALL is the most common childhood malignancy representing nearly one-third of pediatric cancers [32]. Several studies reported the increase in WT1 mRNA to be one of risk factors resulting in relapse in AML [33-35]. Moreover, overexpression of WT1 mRNA was frequently detected in de novo childhood ALL [23,36,37]. We showed high positive rates of WT1 mRNA expression (approximately 90%) in the PB and BM samples of de novo childhood and adult ALL patients. They were also expressed at a high frequency.
In this study, it is considered that there is no difference in WT1 mRNA expression level and positive rate between childhood ALL and adult ALL before treatment, so it is speculated that the kinetics of WT1 mRNA expression level after treatment will show the same trend for both childhood ALL and adult ALL. From the literature of the authors [38-41], WT1 mRNA expression level in adult ALL reflects the therapeutic effect and has been shown to be useful as a monitoring marker.
In Japan, the incidence of ALL by subtype has been reported as 70.5% for B-precursor ALL, 14.0% for pre-B ALL, 2.5% for mature B-ALL, and 13.0% for T-ALL [31]; a similar distribution was also demonstrated in this study. Furthermore, subtype B-precursor ALL showed higher mean values of WT1 mRNA levels than pre-B ALL. This suggests higher levels of WT1 mRNA expression in undifferentiated hematopoietic cells, which is consistent with previous studies that reported higher WT1 mRNA levels in B-precursor ALL than in other subtypes [22,42]. Moreover, intracellular WT1 mRNA levels decreased when K562 cells (a CML-derived cell strain) differentiated into erythroids or megakaryocytes, and when HL60 cells (a promyelocytic leukemia-derived cell strain) differentiated into granulocytes, monocytes, and macrophages via various inducers of differentiation [43,44].
The correlation of WT1 mRNA expression in both, PB and BM, was evaluated in this study, which demonstrated a good correlation with a correlation coefficient (r) of 0.85 in childhood ALL and 0.84 in adult ALL. This result was also reported in myelodysplastic syndrome (MDS) and AML [26,45]. PB and/or BM were chosen for clinical monitoring, depending on the patient’s condition and treatment strategy. Measurement of PB was preferred owing to its convenience and lower burden on the patient.
Fusion gene transcripts were detected in 11 of 49 (22.4%) de novo childhood ALL patients, and the detection rate was similar to that reported in a previous study [16]. Contrastingly, the rate of positivity for WT1 mRNA was 93.8%. Since the levels of the fusion gene transcript and WT1 mRNA correlated well, WT1 mRNA was selected as a monitoring marker instead of the fusion gene transcripts owing to the childhood ALL patient cases that did not express the fusion gene transcripts. The significance was considerable.
Now that a next-generation sequencer has identified cancerous cells to also have multiple clones rather than just one, relapse is thought to occur via subclones that are tolerant to chemotherapy [18].
During the study, a total of 48 childhood ALL patients underwent a follow-up procedure (with the exception of one patient that was transferred), and were assessed to be in remission after starting treatment. During the post-treatment remission phase, WT1 mRNA positivity rates were observed to be 83.3% in both PB and BM, and significantly lower than the rates before treatment (p<0.01; Student’s t-test). To assess hematological remission via ROC analysis, we calculated the cut-off values of WT1 mRNA expression to be 220 copies/μg RNA for PB and 1,820 copies/μg RNA for BM. Although the decrease in WT1 mRNA level after treatment was previously reported in AML and ALL [26,36,46], this study was the first to attempt to obtain the upper cut-off values of WT1 mRNA expression indicating the remission phase. Miyawaki et al. reported that relapse in AML patients was possible if WT1 mRNA levels rose above 200 copies/μg- RNA in PB from the remission phase [20]. Since this value was similar to the PB upper cut-off value obtained in the present study, it seemed appropriate to use it as the reference value for early relapse in ALL. MRD after induction therapy is useful for stratification of the treatment [13]. Optimization of treatment using stratification results in improved prognosis of ALL treatment. Measurement of WT1 mRNA is useful for stratification, which is thought to contribute to the improvement of treatment outcome. Common MRD targets include recurrent cytogenetic abnormalities and mutations in important hematological genes. Unfortunately, well-characterized targets are lacking in many ALL patients. WT1 is expressed in leukemic stem cells and in most cases of AML and ALL.
In this study we performed PCR to detect Ig/TCR gene rearrangements in 6 patients considered to represent severe cases. The results showed that 4 patients were positive and 2 were negative after induction, and the 2 negative patients remained negative after consolidation. In 2 of the 4 positive patients, Ig/TCR gene rearrangement values decreased to 10−4 or less after induction, and became negative after consolidation. For the 2 patients, whose values were 10−4 or more after induction, the values became positive within quantitative range. A clinical study by the AIEOP-BFM group assessing the usefulness of MRD quantification based on Ig/TCR gene rearrangements evaluated by PCR reported that the presence of MRD equal to or greater than 10−3 at time points after consolidation indicates poor prognosis in the patients for whom allogeneic SCT is recommended [11]. This report suggested that our 6 patients also have poor prognosis, which demonstrates the clinical usefulness of the Ig/TCR gene rearrangement evaluation. Meanwhile, the WT1 mRNA levels in the PB of the 6 patients mentioned above were 220 copies/μg RNA or less at all time points, suggesting that the patients achieved remission. Recently the methods of MRD evaluation and interpretation of the evaluation results have been strictly standardized for multiregional clinical trials in Europe, including those for Ig/TCR gene rearrangements [8], which were highly sensitive and useful. As with the Ig/TCR gene rearrangements, the WT1 mRNA levels, also indicating remission, were observed to be reduced. Furthermore, the WT1 mRNA levels indicated remission for not only BM, but also PB, confirming the usefulness of the measurement.
Although 24 out of 48 pediatric patients (50.0%) still expressed ≥ 50 copies/μg RNA of WT1 mRNA in the PB samples after treatment and during the remission phase, the expression levels were observed to have decreased. In these cases, the presence of MRD, which was undetectable in hematological remission, was suspected. Detection is considered difficult when the blast cell is only available in minimum amounts of MRD during microscopic examination. The depth of remission was assumed to be shallower in these cases than in cases that showed a decreased level in PB (below LOD) after treatment. The WT1 mRNA levels increased when the disease stage progressed from the remission to the relapse phase in ALL patients [23,36,38-41,46-48], and the depth of remission proved to be much shallower when the expression levels surpassed the upper cut-off values for remission (i.e., 220 copies/μg RNA for PB and 1,820 copies/μg RNA for BM). This type of result suggests relapse and needs to be noted for treatment (Figure 7). Moreover, the WT1 mRNA levels after transplantation also predict recurrence. The level of WT1 mRNA in PB is also useful for immunization after transplantation (e.g., induction of GVL), thereby alleviating the load on the patient [49].
WT1 mRNA detected in high frequency, even in de novo ALL patients exhibiting evidence of fusion gene transcripts, reflected the treatment effects and depth of remission in ALL patients. Therefore, WT1 mRNA may possibly be a useful monitoring marker of MRD in ALL patients.
This study was funded by Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan. Y.H., Y.Kosaka, K.W., K.Kato, M.I., T.K., S.S., A.W., H.H., Y.Koga, M.Hirayama, T.Nakao, T.H., N.U., K.Ishiyama, K.M., M.Hidaka, K.Kitamura, T.T., Y.U., A.M., K.U., Y.Kanda, K.Koh, and K.H. received research funding from Otsuka to conduct this study. Moreover, T.Naoe and H.S. also received personal fees from Otsuka to conduct this study. Y.M. is employee of Otsuka. K.Imai declares no conflict of interest.
We thank all the participating institutions and physicians, including the authors and their principal investigators, for their support during the study. Furthermore, we thank Ms. Tokuko Okuda for performing the flow cytometry analysis.