Mycobacterial Diseases
Open Access
ISSN: 2161-1068
ISSN: 2161-1068
Research Article - (2023)Volume 13, Issue 1
Tuberculosis (TB) caused by Mycobacterium tuberculosis (M. tb) is the major health problem worldwide. It is a contagious, commonly prevalent disease in developing countries including Pakistan. Poverty in developing countries makes this disease an acute health problem. Almost one third of the world’s population is affected by TB. Pakistan ranks 5th among top Tuberculosis infected countries. Among 100,000 individuals about 280 people are infected with this disease in Pakistan. Vaccination is the way to prevent disease occurrence. Bacillus Calmette-Guerin (BCG) is the only licensed vaccine against TB throughout the world. However, protective immunity developed through this vaccine varies in different parts of the world. Hence, there is need to develop new DNA based anti TB vaccines based on information obtained from Mycobacterium tuberculosis whole genome sequencing, as DNA vaccines are capable to elicit both humoral and cell-mediated immune responses. In the present studies, we selected two Mycobacterium tuberculosis genes Rv0577 and Rv3846 for developing DNA vaccine. These genes were amplified through PCR, confirmed through restriction digestion, ligated in pTZ57R/T vector, transformed in DH5α cells. The clones were confirmed through restriction digestion and sequence analysis. In future, these clones can be used in sub-cloning into mammalian expression vectors and tested as DNA vaccines in mice model.
Restriction; Ligation; Cloning DNA; PCR; Bacillus Calmette-Guerin (BCG)
Mycobacterium tuberculosis (M. tb) is the bacterium responsible for causing Tuberculosis (TB) [1] in humans. TB is a pre-historic disease and earlier it was called as white plague of Europe [2-5]. Every year, about 2 million people died because of this contagious disease [6]. Mycobacterium tuberculosis had infected about one third of the world’s population and about 9 million people were diseased [7]. This infectious disease is the major cause of morbidity and mortality particularly in females and old ones [8-11]. It is one of the three main diseases that are directly related to poverty including AIDS and Malaria [8, 12-15]. Earlier, the TB was considered as pulmonary disease but later it was reported from other body parts as well [16]. Respiratory secretions like sputum [17] and aerosols generated by sneezing [18], breathing [19], laughing and coughing of infected individuals are the frequent ways of spreading this disease [17, 20]. Almost all infected individuals retain live Mycobacterium tuberculosis cells in their body and keeps cells in dormant state due to their cell mediated Immunity. It generally progresses very slow from inactive to active state. Mycobacterium tuberculosis growth rate can be enhanced and proved fatal in patients with compromised immunity like HIV patients [21-23]. Patients infected with HIV could increase the possibility of getting TB approximately from 20 to 200 times [23, 24-27].
In latent TB Infection (LTBI), Mycobacterium tuberculosis resists and survives in host immune system and remains in dormant state as compared to active TB [28]. LTBI is a condition in which people remains unaffected but still there is a chance to develop an infection which indicate that it is a contagious disease [28, 29]. Approximately 90% population suffering from LTBI are not considered TB patients [30]. The rate of fatality is increased in patients that are co-infected with HIV [21, 31]. TB bacteria activates the host immune system by developing a constant condition of a resistant infection [32]. Many deaths of young adults occur because of TB and it is the reason that this disease remains prominent worldwide [33, 34]. In Disability Adjusted Life Years (DALYs) lost, the burden of tuberculosis is greater than 80%, and it is the condition in which early deaths occur rather than infection [35]. In 2004, around 1.7 million people infected by TB, involving 264, 000 infected individuals who were also infected by HIV [26, 36]. The universal cause of deaths caused by TB are unsure, because some countries having high burdens of TB accumulate consistent information on the source of death. The modeling based estimation shows that fatality rates of TB could be falling from 1990s to 2000. Mycobacterium tuberculosis is an acid- fast, cylindrical-shaped microscopic organism [37]. The affinity of Mycobacterium tuberculosis to alveolar walls of lungs is very high therefore it is considered as aerophillus bacteria [38]. In humans Mycobacterium tuberculosis is the only cause of TB and pulmonary TB is caused by it by invading the respiratory tract of host, and about greater than 60% of pulmonary TB is caused by it [39].The common symptoms of pulmonary TB includes hemoptysis, weight loss, fever, and cough for the duration of at least three weeks and nocturnal diaphoresis [40- 42].
DNA vaccines have been very effective against many other diseases such as Respiratory Syncytial Virus (RSV) [43-46], influenza [47- 49], HIV [50, 51], corona virus [52, 53], TB [54] and hepatitis [50, 55]. DNA vaccines make continuing immunity rather than live-attenuated vaccines and there is a no chance of re-activation of microorganism [56]. DNA vaccines depends on state of art of molecular biology techniques and well characterized antigens are used in making these vaccines [57-59]. This new field promised to cure infection like, HIV, TB, cancers and many other diseases. In addition, it is cheap and one of the most safe techniques of vaccination [60]. DNA vaccination has stimulated both the humoral and cell mediated immunity and produces a complete immunogenicity of applicant subunit [58]. DNA vaccination is an efficient approach for the appearance of outsider antigen, thus DNA vaccines provides protection against number of pathogens including Mycobacterium tuberculosis [61].In this study we, amplified Mycobacterium tuberculosis genes RV0577 and Rv3846 using PCR, confirmed through restriction digested and ligated in pTZ57R/T vector and transformed DH5α cells. The clones were confirmed through the restriction digestion and sequence analysis. These cells can be used in future for development of DNA vaccines.
Genes
The two genes Rv0577 and Rv3846 were obtained from BEI Resources VA, USA. These genes were amplified and cloned into a vector ptz57R/T of 2886 bp length. Rv0577 is a Mycobacterium tuberculosis complex-restricted secreted protein involved in the methylglyoxal detoxification pathway that have gene length of 786bp, while Rv3846 is superoxide dismutase having 624 bp gene length and it has a role in the survival of Mycobacterium tuberculosis in macrophages.
Primers
The primers were designed manually with kozak and suitable restriction sites were added. The sequences of primers are given below: RV0577F AGATCTGCCACCATGCCCAAGAAGCGAATACAGGCA
Bgl II kozakRV0577R TCTAGACTATTGCTGCGGTGCGGGCTTCAA
Xba1
RV3846F
AGATCTGCCACCGTGGCCGAATACACCTTGCCAGA
Bgl II
kozak
RV3846R TCTAGATCAGCCGAATATCAACCCCTTGG
XbaI
Amplification of genes and electrophoresis
The selected genes Rv0577 and Rv3846 were amplified through PCR under the following conditions. Pre-denaturation (94 °C for 5 minutes), denaturation (94 °C for 45 sec), annealing (60 °C for 45 sec), extension (72 °C, 1 min and 30 seconds), and final extension (72 °C for 10 minutes). The PCR mixture consisted of 1 µL forward primer, 1 µL reverse primer, 1 µL template DNA, 0.4 µL DNTP (10 mMdNTPs Mix), 10X Taq Buffer, 1.2 µL MgCl2, 0.2 µL Taq DNA Polymerase, 13.2 µL deionized water. Final soaking temperature was 4 °C. PCR product was isolated through agarose gel electrophoresis. One percent agarose gel was prepared through adding agarose (0.48 g) in 400 ml 1X TAE buffer at 93 °C for one minute in an oven and then cooled to 60 °C before pouring in gel casting tray. The combs were placed in the gel casting tray. After 20 minutes, the gel was solidified and the combs were removed and the 1X TAE buffer was poured over the gel. DNA sample was loaded through mixing 5 ul of DNA and 2 µl of Bromophenol blue dye. 5 µl of DNA marker 1KB was also loaded in the well. The gel was run at 80 V and 70 A current. The gel was taken out of the tank and soaked in 0.5 µg/mg Ethidium Bromide (EtBr) solution for staining for 10 min. The bands developed were visualized the bands in gel documentation system (Bio Rad, USA).
DNA ligation
The vector pTZ57R/T (mention brand here) was used in DNA ligation. The ligation mixture consisted of 8 µl 5X Rapid ligation buffer, 6 µL PCR product, 1 µL T/A vector, T/4 DNA ligase, and 4 µL nuclease free water. The mixture was incubated at 22 °C in water bath for overnight and stored at 4 °C.
Competent cells preparation
0.1 M solution was prepared by mixing 1.47 g CaCl₂.2H₂O with 100 mL distilled water. Ampicillin negative agar pates were prepared and streaked by top 10 Esterichia coli stocks, and kept in incubator at 25 °C for 24 hours. Broth media was prepared, picked the colony from plate and transferred it in 2mL broth in test tube, this tube was placed on orbital shaker (Being Scientific Incorporation, USA) at 250 rpm overnight. The next day, 100 mL Ampicillin negative broth in test tube was taken and then transferred the 2 mL broth in freshly prepared 100 mL broth media and kept it on shaker overnight. Observed the growth by taking O.D 400 through spectrophotometer.
Took the 50 mL of two falcon tubes that was ice chilled and transferred the 50 mL culture in each tube near with laminar flow to avoid contamination. Put the falcon tubes on ice for 10 min and centrifuged them at 4 °C at 14800 rpm for the time period of 10 min. Supernatant and pallet was separated and discarded the supernatant. For the removal of all the drops of liquid, tubes were inverted on tissue paper. Took the 10 mL of 0.1M CaCl₂ .2H₂O in the tubes having pellet by placing the tubes in ice. For the dissolution of the whole pallet, solution should be mixed by moving the tubes up and down. Continued mixing of contents for at least 30 min. After that centrifuged it at 14,800 rpm at 4 °C. The same procedure was repeated again and discarded the supernatant and re-suspended in 10 mL 0.1M CaCl₂.2H₂O solution. Again dissolve the pellet by gentle shaking and kept it on ice for 30 min. Centrifuged it again for 10 min at 14800 rpm at 4 °C. In the third and last step, added the 2 mL CaCl₂.2H₂O in the tube having pellet and dissolved the pellet by gentle shaking. While keeping the mixture on ice, pellet was dissolved by gentle shaking. The entire mixture was kept in one falcon tube and then incubate it in ice for the time of 15 min.
Glycerol stock of competent cell
The glycerol stock was made by adding 850 µL of competent cell solution with 150 µL of glycerol. The stock was made in ice chilled micro centrifuge tubes.
Transformation
This process required the chilled and autoclaved things so we stored the eppendorfs and tips on ice. For this purpose, we kept the competent cells and DNA ligated sample separately on ice for at least 30 min for the maintenance of temperature of both the tube in equal rate. Transferred the 5 µL of ligated DNA sample to freshly ice-chilled eppendorf tube and transferred the 100 µL of competent cells to that eppendorf tube and gently mixed them. Stored the tubes on ice for 30 min. During this time period, maintained the temperature of water bath at 42 °C. Put the tubes on floater and kept it on already heated water bath for the time of 90 sec. Quickly transferred the tubes on ice for 1-2 min, freshly prepared broth (LB medium) of 500 µL without antibiotics was added and incubate it at 37 °C by placing it in an incubator and bacteria was recovered. Centrifuge the tubes for 2 min at 14800 rpm. Removed the 300 µL of supernatant and dissolved the remaining 200 µL in pellet. Now transferred the solution to already prepared agar plates having antibiotics. Spread the solution by spreader and store the plates at room temperature for 35-40 min for the absorption of liquid. Inverted the plates and incubate them at 37 °C by placing the tube in incubator for overnight. After one day results were observed.
Preparation of agar plates
For this purpose, 2.8 g agar was dissolved it in 100 mL distilled water in a conical flask, autoclaved it and allowed it to cool because ampicillin is heat sensitive. After that, 100 µL ampicillin was added and poured the medium to petri-dishes, then the plates were allowed to solidify for 20 min.
Selection of clones
The media that was freshly prepared in selective transformed plate, the only one colony was picked from that media and LB medium of 1-5 mL had been introduced with suitable antibiotics (ampicillin; penpbritin). Then incubated it at 37 °C for 12-16 hrs with shaking at 200-250 rpm. The volume of conical flask was at least 4 times the volume of culture. Centrifugation of bacterial culture was done at 14,800 rpm for 2 min at 4 °C to harvest the culture in micro- centrifuge in eppendorfs tubes having volume of about 2 mL and it was done twice. Discarded the supernatant and saved the pellet. The unused portion of culture was stored at 4 °C
Plasmid DNA extraction
The method used for plasmid DNA extraction was Sambrook and Russell [62]. Entire process was carried out at RT.100 µL Alkaline lysis solution I (resuspension solution) was taken and re-suspended the pelleted cells in this solution. Then transferred the suspension of cell to micro-centrifuge tube. By pipetting up and down, bacteria were re-suspended completely. Added the alkaline lysis solution II of 200 µL and mixed it completely by shaking the tubes up and down for 4-6 times to make the solution cleared and viscous. Then stored the tubes on ice for 2-5 min. Then added the alkaline lysis solution III (neutralization solution) of 150 µL and immediately mixed it completely through moving the tubes up and down to keep away from the neighboring precipitation of bacterial cell debris. Centrifuged the bacterial lysate at 4 °C for 5 min in micro-centrifuge tubes to make the pellet that have chromosomal DNA cell debris. Now fresh eppendorf tube was taken and transferred the supernatant to that tube through pipetting while remaining the white precipitates (pellet) in the tube. Two volumes of ethanol were added at RT and precipitated the DNA from solution. Then mixed the solution by vortexing and incubated the mixture for 2 min. Centrifuged the mixture in microfuge to collect the precipitated DNA for 5 min at maximum speed at 4 °C. Now gentle aspiration has been done to remove the supernatant and allowed the eppendorf tubes to stand on paper in inverted position to drain all of the fluid. Any drop that adhered the wall of tube was removed by kim-wipe. Added the 300 µL of 70% ethanol to pellet and inverted the closed tube several time. Centrifugation has been done at maximum speed in microfuge at 4 °C for 2 min to recover the DNA. Removed the supernatant again. Added the 50 µL of de-ionized water in it. Stored the purified plasmid at-20 °C.
Confirmation of clones
Confirmation of clones has been done by restriction digestion and finally by sequence analysis. For this purpose, the restriction enzymes Bgl II and Xba I were used to obtain Rv0577 gene fragments separately. The reaction mixture for first restriction digestion of Rv0577 gene consisted of plasmid DNA 12 µL, Bgl II buffer 12 µL, restriction enzyme Bgl II 1 µL, and deionized water 5 µL. The mixture was incubated for 3 hrs in eppendorf. After first restriction digestion of Rv0577 gene, second digestion was done using restriction enzyme Xba I and Xba I buffer. The reaction mixture consisted of restriction enzyme Xba I (1 µL), 12 µL plasmid DNA, 2 µL Xba I buffer 2 µL and deionized water 5 µL. Restriction digestion was done with an enzyme Bgl II for the confirmation of Rv3846 clone. Rv3846 pTZ57R/T was digested through incubating a mixture of Bgl II buffer (2 µL), 1 µL restriction enzyme Bgl II, 12 µL plasmid DNA and 5 µL deionized water in eppendorf at 37 °C for 3 hours. For Rv3846 pTZ57R/T digestion the reaction mixture consisted of restriction enzyme Bgl II (1 µL), restriction enzyme Xba I (1 µL), 2X Tango Buffer (4 µL), plasmid DNA (12 µL) and deionized water (2 µL). This mixture was incubated in eppendorf at 37C for 3 hours. Restriction digestion solutions were prepared in micro-centrifuge tubes separately. Tubes were labelled and then incubated for at least 3 hrs at 37 °C. The solution was loaded in the wells of gel. Gel was allowed to run in gel electrophoresis and the obtained results were recorded by taking photographs.
Sequencing
The isolated construct (T/A-Rv3846) was sent to Center of Excellence in Molecular Biology (CEMB), University of The Punjab, Pakistan along with its primer set for sequencing.
To control Mycobacterium tuberculosis that is intracellular pathogen, DNA based vaccines have been prepared. Two genes construct of Mycobacterium tuberculosis were made in this study that could be used as DNA vaccines. These genes were made from PCR product in which primers were designed with kozak sequences against M. tb genes. Kozak sequence (GCCACC) reported by kozak in 1987 and is required for the expression of gene in mammalian cells. Kozak sequence increases the binding efficiency of ribosome of gene. By restriction digestion and sequence analysis, clones were confirmed.
Amplification of Mycobacterium tuberculosis genes (Rv0577 and Rv3846) by PCR and gel electrophoresis
Mycobacterium tuberculosis genes were amplified by PCR with restriction sites and kozak sequences to clone into pTZ57R/T vector Figure 1.
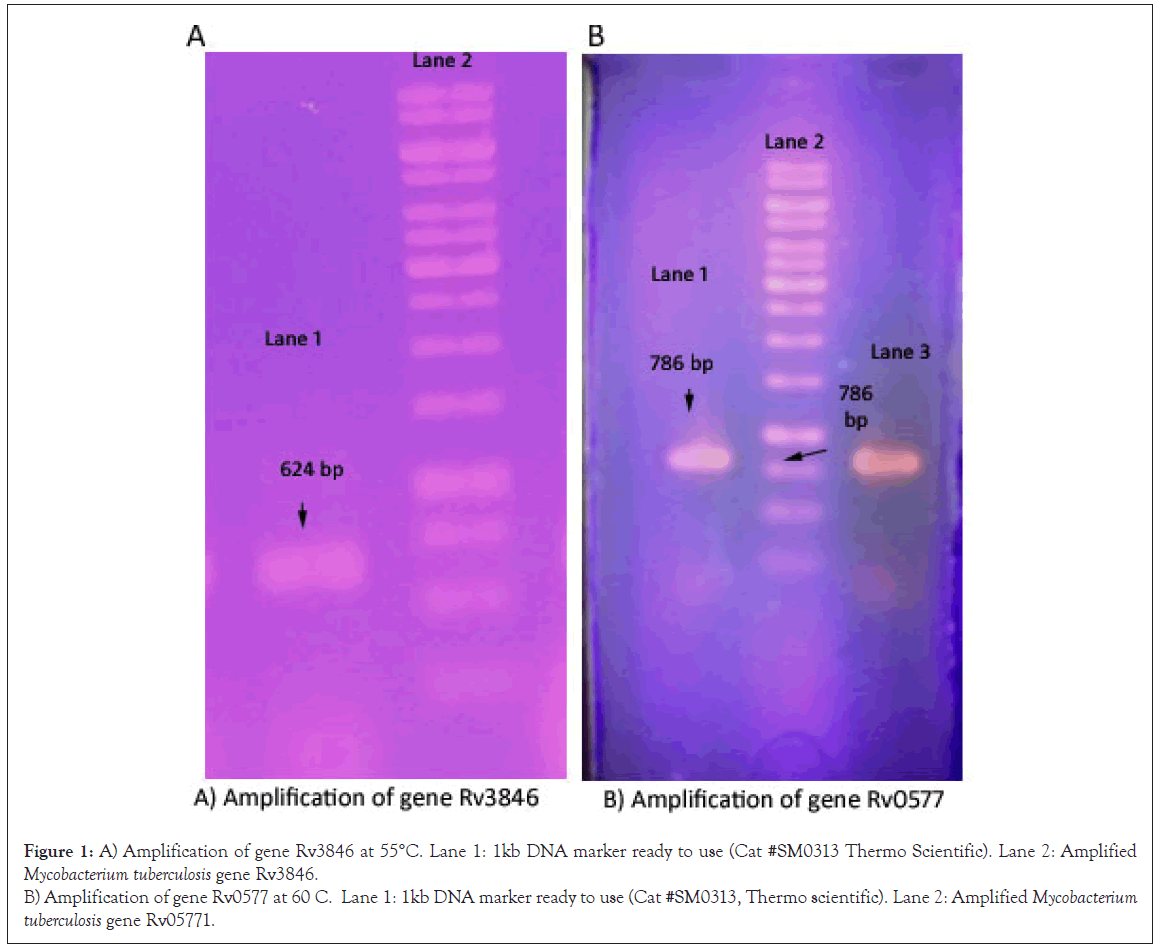
Figure 1: A) Amplification of gene Rv3846 at 55°C. Lane 1: 1kb DNA marker ready to use (Cat #SM0313 Thermo Scientific). Lane 2: Amplified Mycobacterium tuberculosis gene Rv3846.
B) Amplification of gene Rv0577 at 60 C. Lane 1: 1kb DNA marker ready to use (Cat #SM0313, Thermo scientific). Lane 2: Amplified Mycobacterium tuberculosis gene Rv05771.
Transformation
Transformed E.coli cells containing Rv0577 and Rv3846 were grown by just touching the sterilized tips and poured the tip in the broth medium and placed the medium on shaker at 37 °C for 16-18 hrs. Cultures were again grown on LB medium and then the plasmid DNA was extracted from them by alkaline lysis method. Figure 2
Figure 2: Transformed E.coli cells with vector containing genes of interest.
A) Transformed E.coli cells containing gene Rv0577.
B) Transformed E.coli cells containing gene Rv3846.
C) Control.
Confirmation of clones by extracted plasmid DNA
Plasmid DNA was extracted by using alkaline lysis solution. After that through gel electrophoresis, accurate bands of the DNA were obtained Figure 3.
Figure 3: Extracted plasmid DNA.
A) Lane 1: 1kb DNA marker ready to use (Cat #SM0313 Thermo-scientific). Lane 2: Plasmid DNA extract of Rv0577.
B) Lane 1: 1kb DNA marker ready to use (Cat #SM0313 Thermo Scientific). Lane 2: Plasmid DNA extract of Rv3846.
Restriction digestion and confirmation through PCR
Restriction digestion was done for the confirmation of clones (Figure 4 A and 4B). The enzymes XbaI and BglII were used for restriction digestion. The genes Rv0577 and Rv3846 were confirmed through restriction digestion as the band size of 786 bp and 624 bp was obtained after restriction digestion (Figure 4 A and 4B). The restriction product 786 bp and 624 bp genes were further confirmed through PCR and gel electrophoresis (Figure 5 A and 5B).
Figure 4: Restriction digestion of extracted plasmid DNA of gene Rv0577 with Bgl ll and Xba l.
A) Lane 1: Restriction digestion of plasmid DNA with Bgl ll and Xbal. Lane 2: 1kb DNA marker ready to use (Cat #SM0313 Thermo Scientific).
B) Restriction digestion of extracted plasmid DNA of gene Rv3846 with Bgl ll and Xba l, Lane1: Restriction digestion of plasmid DNA with Bgl ll and Xbal. Lane 3: 1kb DNA marker ready to use (Cat#SM0313) Thermo Scientific).
Figure 5: Amplification of gene Rv3846 at 55 °C.
A) Lane 1: Amplified Mycobacterium tuberculosis gene Rv3846. Lane 2: 1kb DNA marker ready to use (Cat #SM0313Thermo Scientific.
B) Amplification of gene Rv0577 at 60 °C. Lane 1: Amplified Mycobacterium tuberculosis gene Rv0577 Lane 2: 1kb DNA marker ready to use (Cat #SM0313 Thermo Scientific).
Sequencing of Rv0577 and Rv3846 cloned into pTZ57R\T vector
Sequencing results indicates that the sequences of both genes were fine and no mutation occur during the process of amplification, cloning and transformation.
Rv0577
ATGCCCAAGAGAAGCGAATACAGGCAAGGCACGCC
GAACTGGGTCGACCTTCAGACCACCGATCAGTCCG
CCGCCAAAAAGTTCTACACATCGTTGTTCGGCTGGG
GTTACGACGACAACCCGGTCCCCGGAGGCGGTGG
GGTCTATTCCATGGCCACGCTGAACGGCGAAGCCG
TGGCCGCCATCGCACCGATGCCCCCGGGTGCACCG
GAGGGGATGCCGCCGATCTGGAACACCTATATCGC
GGTGGACGACGTCGATGCGGTGGTGGACAAGGTGG
TGCCCGGGGGCGGGCAGGTGATGATGCCGGCCTTC
GACATCGGCGATGCCGGCCGGATGTCGTTCATCAC
CGATCCGACCGGCGCTGCCGTGGGCCTATGGCAGG
CCAATCGGCACATCGGAGCGACGTTGGTCAACGAG
ACGGGCACGCTCATCTGGAACGAACTGCTCACGGA
CAAGCCGGATTTGGCGCTAGCGTTCTACGAGGCTG
TGGTTGGCCTCACCCACTCGAGCATGGAGATAGCT
GCGGGCCAGAACTATCGGGTGCTCAAGGCCGGCGA
CGCGGAAGTCGGCGGCTGTATGGAACCGCCGATG
CCCGGCGTGCCGAATCATTGGCACGTCTACTTTGCG
GTGGATGACGCCGACGCCACGGCGGCCAAAGCCGC
CGCAGCGGGCGGCCAGGTCATTGCGGAACCGGCTG
ACATTCCGTCGGTGGGCCGGTTCGCCGTGTTGTCC
GATCCGCAGGGCGCGATCTTCAGTGTGTTGAAGCC
CGCACCGCAGCAATAG
Rv3846
GTGGCCGAATACACCTTGCCAGACCTGGACTGGG
ACTACGGAGCACTGGAACCGCACATCTCGGGTCAGA
TCAACGAGCTTCACCACAGCAAGCACCACGCCAC
CTACGTAAAGGGCGCCAATGACGCCGTCGCCAAACT
CGAAGAGGCGCGCGCCAAGGAAGATCACTCAGCG
ATCTTGCTGAACGAAAAGAATCTAGCTTTCAACCTC
GCCGGCCACGTCAATCACACCATCTGGTGGAAGAA
CCTGTCGCCTAACGGTGGTGACAAGCCCACCGGCG
AACTCGCCGCAGCCATCGCCGACGCGTTCGGTTCG
TTCGACAAGTTCCGTGCGCAGTTCCACGCGGCCGC
TACCACCGTGCAGGGGTCGGGCTGGGCGGCACTG
GGCTGGGACACACTCGGCAACAAGCTGCTGATATTC
CAGGTTTACGACCACCAGACGAACTTCCCGCTAGG
CATTGTTCCGCTGCTGCTGCTCGACATGTGGGAAC
ACGCCTTCTACCTGCAGTACAAGAACGTCAAAGTC
GACTTTGCCAAGGCGTTTTGGAACGTCGTGAACTG
GGCCGATGTGCAGTCACGGTATGCGGCCGCGACC
TCGCAGACCAAGGGGTTGATATTCGGCTGA
In the present studies, we selected the two immune-dominant genes (Rv0577 and Rv3846) and constructed the suitable forward and reverse primers manually for each gene along with kozak sequences (GCCACCATGG). These kozak sequences were well reported sequences as they give extra ribosomal binding sites in eukaryotic immune system [63]. Through PCR amplification of the genes was done by using these primers at suitable annealing temperature. Then ligated the amplified DNA products in a T/A vector pTZ57R/T. The bacteriophage enzyme T4 DNA ligase carried out the process of DNA ligation. The function of T4 DNA ligase is to join short synthetic oligos, two adjacent, plasmid and genomic DNA templates. If a single bp mismatch exists then the efficiency of T4 DNA decreases [64, 65]. Then transformed the ligated products into top 10 strains of E.coli competent cells (DH5α) that are more efficient. CaCl₂ protocol [66] was used for transformation. Then Alkaline lysis solution was used to obtained the high yield of DNA Plasmid. Through PCR and restriction digestion using specific restriction enzymes (Bg1 ll and Xba l), confirmation of DNA plasmid had been done. Finally, plasmid DNA was confirmed through sequence analysis.
Presently, progresses on large scale have been done in the improvement of new TB vaccines, for example inactive whole bacterium, recombinant BCG, attenuated live, DNA vaccine and subunit vaccine [67]. Recently, in adults M. bovis BCG is a single vaccine that is accessible for vaccination against TB. It is observed that defensive efficacies of BCG vaccine are to be relatively variable. Improvement of new efficient vaccines or to improve the BCG efficacy has become essential. In TB vaccine development, recognition of a talented vaccine applicant plays a significant role. Defensive efficiency and immunogenicity of DNA vaccines was compared by individually expressing three significant immune- dominant antigens of Mycobacterium tuberculosis, specifically, ESAT- 6, Ag85B and Ag85A. The study concluded that DNA vaccine may stimulate strong cell-mediated and humoral immunity in mice that is vaccinated and provided additionally a definite degree of defense against Mycobacterium tuberculosis disease. The most excellent defensive efficiency had been rendered via plasmid DNA that encode Ag85A, followed by the plasmid DNA encoding ESAT-6 and Ag85B. The defending efficiency of diverse DNA vaccines had been linked with IFN-g secretion and specific antigen lymphocyte propagation. Though, the same level of protection is not provided by all the three DNA constructs as provided by DNA vaccine that is provided by the BCG vaccine.
The distribution and prevalence of TB is increasing because HIV co-infection morbidity is rising that makes the TB epidemic [68]. Immunity efficacy has been produced by BCG against TB indicated by many reports. But it has failed to stimulate sufficient immunity against pulmonary TB in adults. That is why many researchers like Brandt, et al. [69], Oettinger, et al. [70] and Brandt, et al. [71] realized the significance progress to BCG vaccine and they reported that present vaccine M. bovis BCG is the basis for genetic manipulation for the improvement of its protective efficacy. Experimental studies showed that when BCG vaccines are applied to animals, they immunized successfully and health related problems are also solved. In the world, the existing BCG vaccine has become mainly controversial vaccine that can provide defense only at the age of five in individuals that are vaccinated [72]. DNA based vaccine is the most recent strategy to control the diseases caused by Mycobacterium tuberculosis, an intracellular pathogen. The objectives of current research were to make the clones of Mycobacterium tuberculosis genes (Rv0577 and Rv3846) and to make the DNA vaccine(s) in future use.
In the present studies, we selected the two immune-dominant genes (Rv0577 and Rv3846) and constructed the suitable forward and reverse primers manually for each gene along with kozak sequences (GCCACCATGG). Then amplified these genes through PCR, and ligated the amplified DNA products in a T/A vector pTZ57R/T using T4 DNA ligase enzyme. After that we transformed the ligated products into top 10 strains of E.coli competent cells (DH5α). After that PCR and restriction digestion was done using specific restriction enzymes (Bg1 ll and Xba l). Finally, plasmid DNA was confirmed through sequence analysis. In this experiment we successfully cloned and identified the sequences of Rv0577 and Rv3846 gene. These sequences may be used for the development of vaccines.
In the present studies, we selected two immune-dominant genes Rv0577 and Rv3846 and designed primers. Genes Rv0577 and Rv3846 were successfully amplified and then analyzed them through gel electrophoresis. Chemically treated competent cells were prepared successfully used in transformation studies. The selected clones were confirmed through PCR, restriction digestion and sequence analysis. Then glycerol stock of each positive clone was stored at -80 °C. The commercial sequencing results indicate that the sequences of both genes are fine and no mutation occurs during the process of amplification, cloning and transformation. These immune-dominant genes can be used in future for development of vaccines against tuberculosis disease.
We acknowledge help and support from Mirza Imran Shehzad lab members.
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “conceptualization, M.I.S. and M.Q; methodology, M.Q; software, M.Q; validation, M.Q; and M.I.S; formal analysis, M.Q; investigation, M.I.S; resources and A.H; data curation, M.Q; writing-original draft preparation, M.Q; writing—review and editing, A.H; visualization, A.H; supervision, A.H and M.I.S; project administration, M.I.S; funding acquisition, A.H. All authors have read and agreed to the published version of the manuscript.
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Qasim M, Hameed A, Shehzad MI. (2023) Cloning and Sequencing of Tuberculosis Genes Rv0577 and Rv3846 for DNA Vaccine. Mycobact Dis.13:312.
Received: 02-Jan-2023, Manuscript No. MDTL-23-21732; Editor assigned: 05-Jan-2023, Pre QC No. MDTL-23-21732 (PQ); Reviewed: 19-Jan-2023, QC No. MDTL-23-21732; Revised: 26-Jan-2023, Manuscript No. MDTL-23-21732 (R); Published: 02-Feb-2023 , DOI: 10.35248/2161-1068.23.13.312
Copyright: © 2023 Qasim M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.