Journal of Plant Biochemistry & Physiology
Open Access
ISSN: 2329-9029
ISSN: 2329-9029
Research Article - (2019)Volume 7, Issue 3
Tomato (Solanum lycopersicum Mill.) which belongs to the family Solanaceae is one of the most important vegetable grown and consumed worldwide. Rhodanese activity distribution in the stem, leaf, green unripe fruit, yellow ripening fruit, and red ripe fruit tomato plant parts of tomato plant were compared. The yellow ripening fruit had the highest activity followed by the leaf then the stem then green unripe fruit, while the least activity was shown by the red ripe fruit. The activity difference between the red ripe fruit and those of the stem, leaf and yellow ripening fruit was statistically significant. Also, the activity difference between yellow ripening fruit and red ripe fruit was statistically significant. The purified rhodanese from the almond nuts had a specific activity of 4.45 RU/mg with yield of 0.2%. A Km value of 46.34 mM with Vmax 2.10 RU/ml/min were obtained from KCN while a Km value of 26.34 mM with Vmax of 1.52 RU/ml/min was obtained from N
Tomato, Solanum lycopersicum L., is a member of a diverse family Solanaceae, which have over 3000 species, and they grow in a wide variety of habitats [1]. It is an edible fruit that is grown across the continents. It is consumed raw or cooked, in many dishes, sauces, salads, and drinks. Tomato and its wild relatives are part of a larger monophyletic group (the Potato clade) that also contains the potatoes and their wild relatives [2]. Solanum lycopersicum was previously classified as Lycopersicon esculentum Mill. But due to evidence data from both morphology and molecular sequences in support of its inclusion in the large genus of Solanum L., resulted in its new nomenclature [3-6]. The analyses of multiple data sets from a variety of genes unambiguously establish tomatoes to belong to Solanum [7-12]. Tomato has both nutritive and health benefits. From epidemiological studies, clinical trials and experiments on animals as well as in vitro studies, this protective effect has been mainly attributed to provitamin A and other carotenoids [13]. Carotenoids are important phytochemical compounds that provide precursors to essential vitamins and antioxidants. Because tomato is the second-most important vegetable in the world after potato, with an annual production of around 122.9 million tonnes of fresh weight [14].
Plants are constantly exposed to a wide array of environmental stresses that cause major losses in productivity. Resistance and susceptibility to these biotic and abiotic stresses are complex phenomena, in part because stress may occur at multiple stages of plant development and often more than one stress simultaneously affects the plant. Plants devise a number of physiological and metabolic responses to combat the effect of of the various environmental challenges [15]. Plants as well as other organisms have multi-protein families (MPF) of Sulphurtransferases (Str). Str also called Rhodaneses, comprise a category of enzymes broadly distributed in all phyla, paving the way for the transfer of a sulphur atom from suitable sulphur donors to nucleophilic sulphur acceptors, at least in vitro [16]. Despite the presence of Str activities in many living organisms most especially plants, their physiological role has not been clarified unambiguously [16].
Rhodaneses provide eukaryotic and prokaryotic cells with a labile reactive sulphide which is at the basis of different cellular processes involving sulfur transfer reactions [17,18]. The biological function is still largely debated and spans from cyanide detoxification to sulfur metabolism. The extreme non-determination in attributing a defined role to sulphur transferases (STs) is likely due to the fact that the identification of the in vivo substrates has thus far proven inconclusive [19]. It appears unlikely that privileged natural substrates of Rhodaneses are those identified by in vitro reactions given that the thiosulfate/mercaptopyruvate and cyanide affinity is in the millimolar range, thus apparently incompatible with the supposed physiological role in enzymatic cyanide detoxification [20]. Therefore, alternative functions, including sulfur and selenium metabolism and biosynthesis of prosthetic groups in iron-sulfur proteins, have been proposed [20-25]. So far, the presence of Rhodanese activity has been studied in some plants, animals and microorganisms; its presence has not been studied in tomato plants. This study aims at studying the presence of Rhodanese activity in the stem, leaf, green unripe fruit, yellow ripening fruit, and red ripe fruit tomato plant parts of tomato plant. Furthermore, the purified Rhodanese from the red ripe tomato fruit would be partially characterized.
Materials
Potassium cyanide, sodium thiosulphate, boric acid, sodium borate, formaldehyde, ferric nitrite, nitric acid, citric acid, sodium citrate and ε-amino-n-caproic acid were obtained from BDH Chemical Limited, Poole, England. Glycerol, sodium acetate, sodium dodecyl sulphate (SDS), low molecular weight calibration kit for electrophoresis, ethylenediamine tetraacetic acid (EDTA), Coomassie Brilliant-Blue, Blue Dextran, and Bovine Serum Albumin (BSA) were obtained from Sigma Chemical Company, St. Loius, Mo., USA. Biogel P-100 was purchased from Bio-Rad Laboratories Inc., Benicia Ca., USA. Other chemicals used were of analytical grade and were procured from reputed chemical firms. Tomato plant parts were collected from a local farm at Arada area, Ogbomoso, Nigeria and transported to the laboratory for analysis. They were identified at the Department of Biology, Ladoke Akintola University of Technology, Ogbomoso, Nigeria.
Methods
Enzyme extraction and isolation: In 200 g of the stem, leaf, green unripe fruit, yellow ripening fruit, and red ripe tomato fruit collected were homogenized in three volumes of 100 mM phosphate buffer, pH 6.5 containing 10 mM sodium thiosulphate and with a Warring Blender. The homogenate was filtered through a double layer of cheese cloth and then centrifuged at 4000 rpm at 10°C for 15 min using Centurion cold centrifuge (R-1880). The pellets were discarded and an aliquot of the supernatant was then assayed for Rhodanese activity and protein concentration.
Ammonium sulphate precipitation: In 100 ml of the supernatant of the crude Rhodanese enzyme from the red ripe tomato fruit was brought to 80% ammonium sulphate saturation by slow addition and stirring of 51.6 g solid ammonium sulphate. This was done for 1 h with occasional stirring until all the salt had dissolved completely in the supernatant. The mixture was left for 12 h at 4°C.
Dialysis: The solution from the ammonium sulphate precipitation was centrifuged at about 4,000 rpm for 30 min. The pellet was washed with a small amount of 0.1 M phosphate buffer (pH 7.2) was dialyzed against several changes of 0.1 M solution of phosphate buffer (pH 7.2) at 4°C for 8 hours using a dialysis bag. The dialysate was centrifuged at 4,000 rpm for 15 min and the supernatant was assayed for Rhodanese activity and protein concentration.
Enzyme assay: Rhodanese activity was measured according to the method described by [26]. The reaction mixture consists of 0.25 ml of 50 mM borate buffer (pH 9.4), 0.1 ml of 250 mM KCN, 0.1 ml of 250 mM Na2S2O3 and 0.1 ml of the enzyme solution in a total volume of 0.55 ml. The mixture was incubated at 37°C for 1 min and the reaction was stopped by adding 0.25 ml of 15% formaldehyde, followed by the addition of 0.75 ml of Sorbo reagent. The absorbance was taken at 460 nm.
Protein determination: Protein concentration was determined by the method of Bradford using Bovine Serum Albumin (BSA) as the standard, where the protein absorbance was interpolated from a standard protein curve. The reaction mixture consists of 10 μl of the enzyme solution and 1.0 ml of Bradford reagent. The absorbance was taken at 595 nm.
Ion-exchange chromatography on CM-Sephadex C-50: CMSephadex C-50 cation exchanger was pretreated by boiling five grams (5 g) of the resin in distilled water for 1 h. This was followed by the addition of 100 ml 0.1 M HCl for 30 min, after which the acid was decanted and the resin was washed with distilled water several times to ensure the total removal of the acid. Thereafter 100 ml of 0.1 M NaOH was added to the resin, which was decanted after 30 min, followed by the thorough rinsing of the resin with distilled water to remove all traces of the base. The resin was then equilibrated with 0.1 M phosphate buffer (pH 7.6) before it was packed into a 2.5 x 40 cm column. Six milliliters (6 ml) of the enzyme solution from the preceding step was then applied on the column. The column was washed with 0.1 M phosphate buffer (pH 7.6) to remove unbound protein, followed by a step-wise elution with 0.5 m NaCl and 1.0 M NaCl in the same buffer. Fractions of 4 ml were collected from the column at a rate of 48 ml per h. The active fractions from the column were pooled and dialyzed against 50% glycerol in 0.1 M phosphate buffer, pH 7.6. The dialyzed fraction was assayed for Rhodanese activity and protein determination using Bradford reagent, the active fractions pooled were stored in a refrigerator.
Size exclusion chromatography on Biogel P-100 and determination of native molecular weight: Three milliliters (3 ml) of enzyme solution from the preceding purification step was then layered on the biogel P-100 column. Fractions of 2 ml were collected from the column at a rate of 12 ml per h. The active fractions from the column were pooled and dialyzed against 100 ml 50% glycerol in 0.1 M phosphate buffer, pH 7.5. The dialyzed fraction was assayed for Rhodanese activity and protein.
The native molecular weight was determined on a biogel P-100 (2.5×90 cm) column. The standard proteins were bovine serum albumin (Mr 66000; 5 mg/ml), ovalbumin (Mr 45000; 5 mg/ml) and chymotrypsinogen-a (Mr 25,000; 5mg/ml). Total sample volume of each of the protein markers applied to the column was 5 ml. The proteins were eluted with phosphate buffered saline pH 7.2. Fractions of 5 ml were collected and monitored by measuring absorbance at 280 nm for the protein. The void volume (V0) of the column was determined by the elution volume (Ve) of Blue dextran (2 mg/ml). A 5 ml aliquot of the enzyme solution was then applied to the same column and the elution volume of the Rhodanese was determined.
Determination of kinetic parameters: The kinetic parameters (Km and Vmax) of the enzyme were determined. The Km and Vmax of sodium thiosulphate and potassium cyanide were determined by varying concentrations of sodium thiosuphate and potassium cyanide between 10 mM and 50 mM at a fixed concentration of 25 mM potassium cyanide and sodium thiosuphate respectively in 50 mM buffer pH 9.4. The kinetic parameters were determined from the double reciprocal plot.
Effect of pH on the Rhodanese activity: The effect of pH was studied by assaying the enzyme using different buffers: 0.1 M citrate buffer (pH 3.0-6.0); 0.1 M phosphate buffer (pH 7.0-8.0) and borate buffer (pH 9.0-10.0). The reaction mixture of 1.0 mL contained 0.25 mL of buffer and other reagents as contained in the normal Rhodanese assay protocol.
Effect of temperature on the Rhodanese activity: The enzyme was assayed at temperatures between 30°C and 100°C to investigate the effect of temperature on the activity of the enzyme and to determine the optimum temperature of the enzyme. The assay mixture was first incubated at the indicated temperature for 10 min before initiating reaction by the addition of an aliquot of the enzyme which had been equilibrated at the same temperature. The Rhodanese activity was assayed routinely as previously described.
Effect of salts on the enzyme activity: The salts tested were NaCl, KCl, BaCl2, HgCl2 and MnCl2 at 0.001 mM, and 0.01 mM from stock solutions of 0.1 mM in a typical Rhodanese assay mixture. The metallic chlorides were dissolved in distilled water. The reaction mixture without the salts was taken as control with 100% activity.
Substrate specificity: The substrate specificity of the enzyme was investigated by testing its activity towards structurally similar compounds. The compounds include Mercapto-ethanol (MCPE), Ammonium per sulfate (NH4)2S2O8), Ammonium sulfate (NH4)2SO4), Sodium sulfite (Na2SO4), Sodium metabisulfate (Na2S2O5). The solutions of the compound (250 mM) were prepared in 50 mM borate buffer, pH 9.4 and assayed as described above.
Rhodanese activity distribution in tomato plant parts
The mean difference between the red ripe tomato fruit and those of the stem, leaf and yellow ripening tomato fruit was statistically significant. Also, the mean difference between yellow ripening fruit and red ripe fruit was statistically significant (Table 1).
| Tomato parts | Rhodanese Activity (RU/ml/min) |
|---|---|
| Stem | 0.34 ± 0.012 |
| leaf | 0.36 ± 0.012 |
| green unripe fruit | 0.33 ± 0.012 |
| yellow ripening fruit | 0.39 ± 0.005 c |
| red ripe fruit | 0.27 ± 0.018 a,b,d |
Table 1: Comparison of Rhodanese activity distribution in tomato plant parts.
Purification
The result of the purification protocol for the red ripe tomato fruit Rhodanese showed a specific activity 4.45 RU/mg with yield of 0.2%. The purification procedure is summarized in (Table 2) and (Figures 1 and 2) shows elution profile of Ion-exchange chromatography on CM-Sephadex C-50 and Biogel P-100 size exclusion chromatography respectively.
| Total Protein (mg) | Total Activity (U) | Specific Activity (u/mg) | Yield% | Purification Fold | |
|---|---|---|---|---|---|
| Crude Enzyme | 140.78 | 286.55 | 2.04 | 1 | 1 |
| 80% (NH4)2SO4 Precipitation | 53.9 | 150.63 | 2.79 | 0.53 | 1.37 |
| CM-Sephadex C-50 Ion Exchange Chromatography | 31.45 | 94.54 | 3.01 | 0.33 | 1.48 |
| Biogel P-100 Size Exclusion Chromatography | 12.62 | 56.1 | 4.45 | 0.2 | 2.18 |
Table 2: The purification profile of Rhodanese from the tomato fruit.
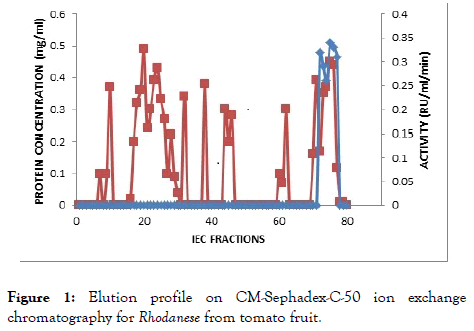
Figure 1. Elution profile on CM-Sephadex-C-50 ion exchange chromatography for Rhodanese from tomato fruit.
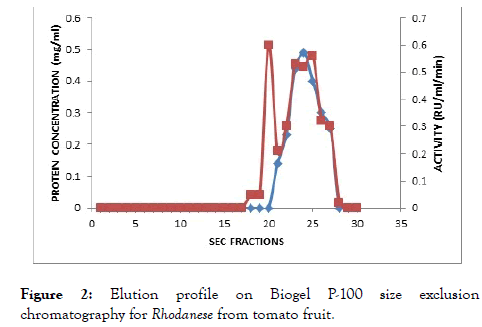
Figure 2. Elution profile on Biogel P-100 size exclusion chromatography for Rhodanese from tomato fruit.
Kinetic parameter
Rhodanese followed Michaelis-Menten kinetics with a Km for sodium thiosulphate (Na2S2O3) of 46.34 mM with Vmax of 2.10 RU/ml/min estimated from a double reciprocal plot (Table 3 and Figure 3). The Km for potassium cyanide (KCN) of 26.34 mM with Vmax 1.52 RU/ml/min estimated from double reciprocal plot (Figure 4).
| Substrate | Km | Vmax |
|---|---|---|
| Potassium cyanide | 46.34 | 2.1 |
| Sodium thiosulphate | 26.34 | 1.52 |
Table 3: Showing the kinetic values of the Potassium cyanide and Potassium cyanide substrates.
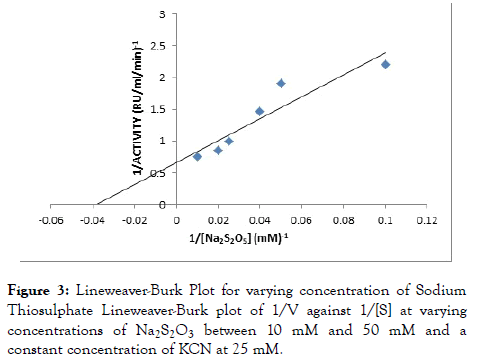
Figure 3. Lineweaver-Burk Plot for varying concentration of Sodium Thiosulphate Lineweaver-Burk plot of 1/V against 1/[S] at varying concentrations of Na2S2O3 between 10 mM and 50 mM and a constant concentration of KCN at 25 mM.
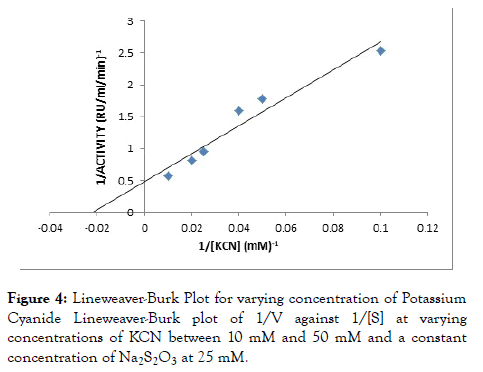
Figure 4. Lineweaver-Burk Plot for varying concentration of Potassium Cyanide Lineweaver-Burk plot of 1/V against 1/[S] at varying concentrations of KCN between 10 mM and 50 mM and a constant concentration of Na2S2O3 at 25 mM.
Effect of temperature
The effect of temperature on the enzyme was determined by assaying at different temperatures between 30°C to 100°C to investigate the effect of temperature on the enzyme activity. The optimum activity was obtained at 45°C as reflected on below (Figure 5).
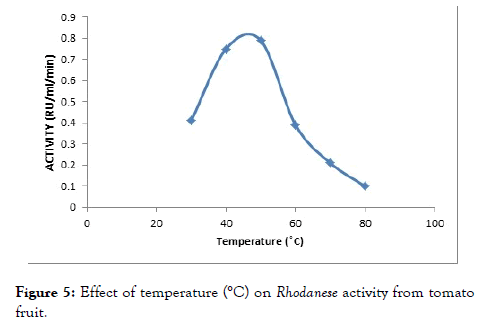
Figure 5. Effect of temperature (°C) on Rhodanese activity from tomato fruit.
Effect of pH
The effect of pH on the enzyme was determined using assay buffer ranging from 3 to 11. The pH assay is done to determine the optimum pH of enzyme activity. An optimum pH of 8.0 was observed (Figure 6).
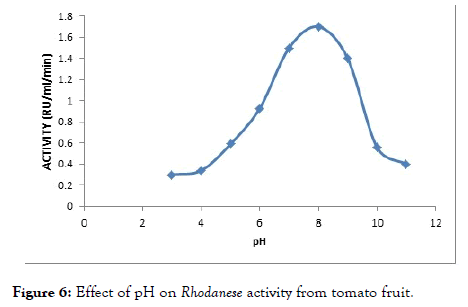
Figure 6. Effect of pH on Rhodanese activity from tomato fruit.
Substrate specificity
Substrate specificity of Rhodanese was investigated by testing its activity towards structurally related sulfur compounds in a typical Rhodanese assay mixture. The activity was expressed as a percentage activity of the enzyme using sodium thiosulphate as the control (Table 4).
| Substrate | %Activity |
|---|---|
| Sodium thiosulphate | 100 |
| Sodium Metabisulphite | 37.45 |
| Ammonium persulphate | 57.11 |
| Mecaptoethanol | 59.9 |
Table 4: Showing the percentage substrate specificity of each sulphur compound.
Effect of metals
The effect of metal ions (NaCl, KCl, HgCl2, BaCl2, MnCl2) at concentrations of 1 mM and 10 mM using their chloride salts was studied. The enzyme activity was inhibited by HgCl2, MnCl2 and BaCl2 in a concentration dependent manner while KCl, CaCl and NaCl had no pronounce effect on the enzyme as shown in the (Table 5).
| Chloride Salts | %Residual Activity (1 mM) | %Residual Activity (10 mM) |
|---|---|---|
| HgCl2 | 75 | 67 |
| KCl | 95 | 93 |
| CaCl2 | 84 | 79 |
| NaCl | 96 | 94 |
| BaCl2 | 85 | 70 |
| MnCl2 | 78 | 66 |
Table 5: Effect of metals on Rhodanese from ripe tomato fruit.
The presence of Rhodanese activity has been studied from various sources, such as Bacillus cereus. Pentadiplandra brazzana, a ginger rhizome, almond nuts tapioca leaf and goat liver [27-31]. This study evaluated and compared the distribution of Rhodanese activity in the stem, leaf, green unripe fruit, yellow ripening fruit, and red ripe fruit tomato plant parts (Table 1). The yellow ripening fruit had the highest activity followed by the leaf then the stem then green unripe fruit, while the least activity was shown by the red ripe fruit. The mean difference between the red ripe fruit and those of the stem, leaf and yellow ripening fruit was statistically significant. Also, the mean difference between yellow ripening fruit and red ripe fruit was statistically significant. The increase in Rhodanese activity in the yellow ripening fruit could be as a result of endogenous cyanide release during ripening and senescence of the tomato fruit as compared to the green and red ripe fruit. During ripening and senescence of fruit, there is conversion of 1-amino-cyclopropane-1-carboxylic acid to ethylene which liberates cyanide in equimolar amounts as ethylene [32]. Possibly due to the functional role the leaf as a major site energy transformation, hence, its high activity of Rhodanese as compared to the stem. Rhodanese activity has been detected in several tissues parts of animals such as the liver, proventriculus, esophagus, gizzard, cecum, brain, large intestine, duodenum, crop, spleen, trachea, pancreas, heart, kidney, lung [33-41].
In plants Rhodanese activity has been observed in the chloroplasts from several plants and its activity correlate with the labile sulphide concentration in the plant [42]. Rhodanese has been purified from cabbage leaves and shown to be able to reactivate ferredoxin from apoferredoxin [43-45]. Investigated and detected the expression of Rhodanese in crude plant extracts of nine randomly selected plant tubers namely, sweet potato (Ipomoea batatas), yellow yam, Irish potato (Solanum tuberosum), bitter yam (Diascorea bulbifera), cocoyam, sweet yam (Diascorea esculentu), water yam (Diascorea alata) and cassava (Manihot esculentu).
Rhodanese from the red ripe tomato fruit was further purified by ammonium sulphate precipitation, CM-Sephadex C-50 Ion Exchange Chromatography and size exclusion chromatography on biogel-P 100 and partially characterized. It had a specific activity of 4.45 RU per mg of protein and 0.2% recovery (Table 2). Different purification values have been reported by other researchers in their work. 27. Itakorode et al. [27] reported a purification yield of 36.8% and a specific activity of 25.30 μmol/min/mg for Rhodanese extracted from Bacillus cereus. 28. Okonji et al. obtained a value of specific activity of 4.82 RU/ mg of protein with 19.8% yield from the root of Pentadiplandra brazzeana. A yield of 7.8 was obtained in tapioca leave [46] and a specific activity of 5.4 RU per mg of protein. Ehigie et al. [47] reported a purification yield of 9.06% and specific activity of 0.47 RU/mg for Zinginber officinale and had a specific activity of 5.09 RU/mg with yield of 0.06% almond nuts.
Researchers on Rhodanese have reported various affinities between the substrates to the active site of the enzyme. The red ripe tomato fruit Rhodanese had a higher affinity for sodium thiosulphate compared to potassium cyanide. This is in tanderm with the affinities demonstrated by Rhodanese from the root of Pentadiplandra brazzeana, Zinginber officinale rhizome, tapioca leaf, Bacillus cereus and goat liver [27,29-31]. The apparent Km values from this study for KCN and Na2S2O3 were 46.34 and 26.34 mM, respectively (Table 3). However, contrasting higher affinity for potassium cyanide compared to sodium thiosulphate has been reported. Such studies include that of Rhodanese from almond leaf, fruit bat liver, hepatopancreas of Limicolaria flammea, and guinea pig kidneys [28,29,40]. By implication, experiments show that the catalytic mechanism of Rhodanese is a non-sequential double displacement catalytic mechanism. Either potassium cyanide or sodium thiosulphate may bind first to the Rhodanese active site [48-50].
The substrate specificity study on Rhodanese from the tomato has preference for sodium thiosulphate as substrate compared to the other substrates (Table 4), which is in line with the findings reported [27-29,50] was the first to study the ability of different thiosulphates to substitute thiosulphate in Rhodanese reaction. Cyanide and thiosulphate were reported to be acceptor substrates, while sulphite, persulphide and sulphinates were reported to serve as donor substrates [51].
The effect of metals on Rhodanese from tomato showed that at 1 mM concentration the metals used relatively did not affect the activity of the enzyme. At 10 mM concentration, the divalent metals; Mncl2 Hgcl2, Cacl2, Bacl2 relatively inhibited the enzyme (Table 5). This may be due to induction changes in the conformation of the enzyme or the interaction of these metal ions with sulphydryl groups at the enzyme catalytic site [52,53].
Rhodanese from tomato fruit showed maximum activity at pH 8.0 (Figure 5). This is in tanderm with optimunn pH obtained in Mudskipper liver, and Pentadiplandra brazzeana (Baill) root [28,54], Generally, the optimum pH range of 7.0-11.0 have been reported by researchers. An optimum pH of 9.0 was reported for ginger, almond and Bacillus cereus Rhodanese respectively. Chew and Boey in 1972 worked on tapioca leaf and obtained a value of 10.2 to 11.
An optimum temperature of 45°C was obtained for the tomato fruit Rhodanese (Figure 6). Akinsiku et al. [55] reported an optimum temperature of 40°C for catfish liver Rhodanese. It was reported an optimum temperature of 50°C for ginger, almond, bail root, bovine liver Rhodanese and mudskipper liver Rhodanese respectively. Okonji et al. obtained a wide optimum temperature of 60°C for Bail root Rhodanese [26,28,49,54]. The high temperature might be as a result of adaptation to harsh environmental condition.
This study shows the distributive presence of Rhodanese activity the stem, leaf, green unripe fruit, yellow ripening fruit, and red ripe fruit tomato plant parts. Further, purification and characterization of the red ripe tomato fruit showed properties peculiar to Rhodanese from other sources. The presence of the activity of Rhodanese in the samples suggest its possible role in other physiological activities apart from cyanide detoxification. Further study into the Rhodanese protein may be exploited in bioremediation of cyanide polluted soil.
Citation: Ehigie AF, Abdulrasak MA, Adeleke GE, Ehigie OL (2019) Comparison of Rhodanese Activity and Distribution in Tomato (Solanum lycopersicum Mill.) Plant Parts and its Physicochemical Characterization. J Plant Biochem Physiol. 7:240.
Received: 01-Jun-2019 Accepted: 02-Jul-2019 Published: 09-Jul-2019
Copyright: © 2019 Ehigie AF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.