Journal of Physical Chemistry & Biophysics
Open Access
ISSN: 2161-0398
ISSN: 2161-0398
Research Article - (2015) Volume 5, Issue 1
Cisplatin is an important anti-cancer agent widely used in the clinic; however, it has several notable limitations. To develop novel platinum analogues, key characteristics were considered that may result in more effective platinum analogues. In this study, we present comprehensive molecular dynamics simulation studies using a 12-mer DNA (5’-CCTCTggTCTCC-3’, gg= the site of platination) oligonucleotide which was platinated with cisplatin (1), oxaliplatin_1R_2R (2), and BNP3029 (3, a novel substituted cyano platinum analogue, PtCl2[N≡C(CH2)3(C6H5)]2), and analyzed the large data output using the Kolmogorov-Smirnov statistical analyses. In summary, data indicated that BNP3029-DNA had less A-like DNA morphology in comparison to cisplatin-DNA and oxaliplatin-DNA thus maintaining a more B-like DNA form. BNP3029 demonstrated more potent cytotoxic activity, relative to cisplatin and oxaliplatin, in a variety of human cancer cell lines, including several platinum-resistant cell lines.
<Keywords: Cisplatin; Oxaliplatin; BNP3029; Anti-cancer agents
For more than 35 years, cisplatin has been an effective anticancer agent against a variety of tumors including germ cell tumors, ovarian and bladder carcinomas, squamous cell tumors of the head and neck, esophageal cancers, and non-small cell lung tumors either as a single agent or in combination with other chemotherapy drugs (Figure 1) [1-3]. But cisplatin has clinically important limitations including nephrotoxicity, neurotoxicity, nausea, and ototoxicity. Additionally, tumors can develop platinum resistance rendering the drug ineffective [1]. Analogues of platinum such as carboplatin and oxaliplatin [4] (Figure 1) have been developed and approved, but their use is limited compared to cisplatin. Similarly, Nedaplatin, Heptaplatin and Lobaplatin are later generation platinum analogues that have limited country-specific approval [3] (Figure 1).
The postulated mechanism of action of cisplatin involves the initial replacement of one of the two chlorine atoms in cisplatin by water intracellularly, resulting in the formation of a reactive monoaquomonochloro species [3]. This reactive monoaquo species attacks the exposed imidazole N7 atom of guanine on DNA, initially yielding a mono-platinum-DNA adduct. Once this platinum-DNA adduct is formed, the second chlorine atom on cisplatin is displaced; this species then attacks N7 atom on an adjacent guanine forming a 1,2-GG intrastrand adduct with DNA. In the majority of cases, cisplatin forms an intrastrand DNA adduct and has a well documented selectivity for adjacent GG dinucleotide sequences [1]. The 1,2- platinum-DNA adducts have the potential to be cytotoxic and/or induce apoptosis by the inhibition/modulation of multiple intracellular pathways important for cell growth, cell division and cell survival, including replication and transcription. However, some patients develop resistance to cisplatin due to: (i) increased drug efflux from the cell; (ii) decreased accumulation of the drug; (iii) increased inactivation of cisplatin by thiol containing proteins or by glutathione (GSH); (iv) increased repair of the platinum-DNA adducts by nucleotide excision repair (NER) proteins; (v) defects in mismatch repair which may lead to reduced apoptotic activity upon treatment with platinum agents; and (vi) increased replicative bypass (or trans- lesion synthesis).
In order to develop platinum analogues that overcome the aforementioned limitations, key characteristics were evaluated in studies reported herein, and a series of platinum analogues with substituted cyano groups as the carrier ligand were synthesized [5]. In an earlier study, ab initio computational studies on cisplatin, oxaliplatin and BNP3029 (Figure 1), a representative substituted cyano ligand from our novel platinum complexes [6], were reported. Herein, we present a comprehensive comparative computational study on the geometrical and morphological changes induced by cisplatin, oxaliplatin and BNP3029 upon binding to a 12-mer DNA oligonucleotide using molecular dynamics simulations with key findings described using Kolmogorov-Smirnov based statistical analyses [7].
Molecular dynamics (MD) simulation: B-DNA and platinated- DNA with cisplatin, oxaliplatin and BNP3029
MD simulations were carried out using the AMBER6.0 suite of programs [8] employing the parm99 forcefield. For a better description of α/γ backbone angle conformers in DNA, the parm99 forcefield was modified with the inclusion of parmbsc0 forcefield [9]. The 5’-TCTCCGGTCTCC-3’ DNA sequence for B-DNA was used for each MD run. In the case of platinated-DNA, N7 atoms of the two guanines (gg) attach to platinum atom forming the platinum-DNA adduct (5’-TCTCCggTCTCC-3’). Published parameters for terms involving platinum with DNA bases (such as bond, angle and dihedral) were employed [10]. The only deviation from the published parameters was the use of 1.8 Å vdW radius for platinum in the current study based on the recommendation of smaller radius for platinum [11] compared to the 2.2 Å radius used in reference 10.
Total number of MD runs
For B-DNA, five separate runs were performed starting from the same topology and coordinates for the randomly generated initial velocities by using five different seeds. For four of the seeds, MD runs were performed for a period of 10 ns and the last seed was run for 60 ns; thus providing a combined total MD run of 100 ns. For each of the platinated-DNA adducts (with cisplatin, oxaliplatin and BNP3029), five separate runs were performed starting from the same topology (using the electrostatic potential [ESP] charges on L2Pt(1,2-gg) adduct) and coordinates for the randomly generated initial velocities by using five different seeds. Again, for four of the seeds, MD runs were performed for a period of 10 ns and the last seed was run for 60 ns; thus providing a combined total MD run of 100 ns for each of the platinated-DNA adducts. For each of the platinated-DNA adducts (with cisplatin, oxaliplatin and BNP3029), five separate runs were performed starting from the same topology (this time using the restrained electrostatic potential [RESP] charges on L2Pt(1,2-gg) adduct, Figure 1S and see supplementary material for details on RESP charge generation and MD protocol) and coordinates for the randomly generated initial velocities by using five different seeds. For three of the seeds, MD runs were performed for 10 ns and for the remaining two seeds, MD runs were performed for 60 ns; thus providing a combined total MD run of 150 ns for cisplatin-DNA and oxaliplatin-DNA adducts. For BNP3029- DNA, the combined total MD run was 140 ns. In all, a combined total length of 840 ns of MD simulations were performed: platinated-DNA with RESP charges (2*150+1*140=440 ns)+platinated-DNA with ESP charges (3*100=300 ns) + B-DNA (100 ns)=840 ns.
DNA topology data analysis: Subsampling
The MD time series were subsampled according to a measure of their statistical inefficiency, g, so as to produce a set of uncorrelated samples. Based upon the normalized auto-correlation function, R(Δt), (obtained using the solute root-mean square displacement, rmsd, timecourse)
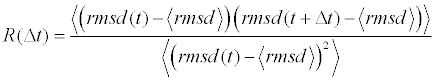
the auto-correlation length τ was computed by direct summation of autocorrelation function:
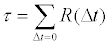
The summation was carried out until R(Δt) falls below 0.05. The statistical inefficiency was then taken as g=1 + 2τ.
Statistical analyses
For purposes of statistical analysis, sets of uncorrelated samples of DNA morphological parameters were obtained by setting the Curves+ [12] stride to g, the statistical inefficiency described earlier. Prior to statistical analysis, the sugar parameter values output by Curves+ were recast into the appropriate 2π interval so as to yield continuous distributions. Descriptive statistics as well as Kolmogorov-Smirnov (KS) statistics were obtained using version 2.12.1 of the R package [13].
Analysis of the morphological changes in platinated-DNA (5’-CCTCTggTCTCC-3’) using the ‘T3 C4 T5 g6 g7 T8 C9 T10’ bases
DNA morphology is conventionally divided into: intra base-pair parameters [Shear (Sx), Stretch (Sy), Stagger (Sz), Buckle (κ), Propel (ω), and Opening (σ)]; base-pair axis parameters [Xdisp (dx), Ydisp (dy), Inclination (η), and Tip (θ)]; inter base-pair parameters or basepair step parameters [Shift (Dx), Slide (Dy), Rise (Dz), Tilt (τ), Roll (ρ), Twist (Ω), Helical-Rise, and Helical-Twist. The first six parameters correspond to transformation between successive base pairs. Helical- Rise and Helical-Twist correspond to the translation and rotation of successive base pairs along and around the helical axis]. Backbone parameters [alpha (α), beta (β), gamma (γ), delta (δ), epsilon (ε), zeta (ζ), chi (χ), phase (P), and amplitude (τmax, the sugar puckering amplitude measures the extent of deviations of the five torsion angles in the five-membered ring from zero)] for each strand (W-the strand that has the platinum adduct; C- the complementary strand); minor and major groove depths and widths; total bend (tbend) angle of DNA; and Axis-bend, which is a re-rendering of total bend using the local base-step data, for a total of 42 parameters.
Other data presented herein include sugar pucker distribution in A-form (combining C3’-endo, C4’-exo and C2’-exo sugar puckers) or in B-form (combining C2’-endo, C3’-exo, C1’-exo and O4’-endo sugar puckers ); % distribution of BI and BII forms of DNA based on the backbone angles ε and ζ (DNA is classified as BI if the difference between the backbone angles ε-ζ <0, and as BII if ε-ζ >0); and % distribution of α and γ backbone angles in the g- / g+ conformations.
The DNA morphological parameters are presented as heat-maps. For B-DNA and for each platinated-DNA adduct, all of the uncorrelated snapshots from the five separate MD runs were combined to obtain the morphological parameters using Curves+ for eight bases (T3 to T10) excluding the two end bases from either side of the platinum adduct. Uncorrelated snapshots for platinated-DNA adduct were combined separately for runs performed using ESP (100 ns) or RESP (150 or 140 ns) charges. Accordingly, for each platinated-DNA adduct, two heat maps are generated: one from the runs using the ESP charges and the other from the runs using the RESP charges. The heat-map is presented with four colors: (i) background light pink- the color refers to Kolmogorov-Smirnov (KS) deviation for a given parameter from that of normal B-DNA in the range of 0 ≤ p<0.8 (p is the probability deviation, as p gets closer to 1, then higher the probability of deviation); (ii) dark yellow- the color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.8 ≤ p<0.9; (iii) orangethe color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.9 ≤ p<0.95; and (iv) brown- the color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.95 ≤ p<1.0. For each of the aforementioned 42 morphological parameters, each base (T3 to T10) contains five cells representing the five MD runs (either 10 ns or 60 ns). A given morphological parameter, for a given base in platinated-DNA adduct, is assigned as significantly different compared to the same parameter for the same base in B-DNA if at least four or more cells are brown in color.
Molecular dynamics studies on platinum-DNA adducts
Platinum-DNA oligonucleotide adducts have been studied using crystallography [14-16], NMR [17,18], and computational approaches [19-21]. Crystallographic studies have previously been carried out on 12-mer DNA oligonucleotide adducts of cisplatin, oxaliplatin and JM118 (JM118 is the active metabolite of Satraplatin) by Lippard and coworkers (14-16). Studies from cisplatin-DNA crystal structure indicated that: (a) platinum coordination bends the DNA (12-mer) by 35-40° towards major groove; (b) DNA takes on A-form morphology on the 5’-side of the platinum adduct with a widened minor groove and adopts B-form morphology on the 3’-side of the platinum adduct; (c) the two guanines which are coordinated to platinum (g6g7, the numbering is based on the 12-mer DNA sequence: 5’-CCTCTggTCTCC-3’) have a roll angle of 26°; (d) the 5’-NH3 hydrogen of cisplatin is in hydrogenbond contact with the g6 phosphate backbone oxygen atom [14]. Similar observations (points: a-d) along with an additional hydrogenbond contact between 3’-NH2 hydrogen of JM118 with O6 atom of g7 are also noted in the crystal structure of JM118-DNA [15]. In the case of oxaliplatin-DNA crystal structure, along with points: a-c as mentioned above, the 3’-NH2 hydrogen of DACH is in hydrogen-bond contact with the O6 atom of g7 as seen in JM118-DNA crystal structure [16] and 5’-NH2 hydrogen of DACH did not have any hydrogenbond contacts with the phosphate backbone as seen with the 5'-NH3 hydrogens of cisplatin-DNA.
The solution studies on platinum-DNA adducts using NMR methods presented different set of numbers compared to the crystal data for some of the parameters mentioned above. The NMR structure of cisplatin-DNA (pdb code: 1A84) [17] showed (i) considerable roll angle (44.7°, Curves+ evaluation) between the two guanines (g6g7) compared to the x-ray structure; and (ii) a significant total bend angle of 83.2° for DNA compared to the x-ray structure. The following contacts observed in the NMR study are not seen in the x-ray structure of cisplatin-DNA: (i) contacts between 5’-NH3 hydrogen with O6 atom of g6 (H---O: 2.786 Å); (ii) contacts between 5’-NH3 hydrogen with the O4 atom of T5 (H---O: 2.875 Å); (iii) contacts between 3’-NH3 hydrogen and the O6 atom of g7 (H---O: 3.595 Å); and (iv) contacts between 3’-NH3 hydrogen of cisplatin and the O4 atom of T8 (H---O: 3.373 Å).
The NMR structure of oxaliplatin-DNA (pdb code: 2K0U) also showed: (i) considerable roll angle (52.6°;, Curves+ evaluation) between the two guanines (g6g7) compared to the x-ray structure; (ii) a total bend angle of 35.6°; for DNA; and (iii) contacts between 3’-NH3 hydrogen and O6 atom of g7 (H---O: 3.026 Å). The following contacts observed in the NMR study are not seen in the x-ray structure of oxaliplatin- DNA: (i) hydrogen-bond contacts between 5’-NH2 hydrogen of DACH with g6 phosphate backbone oxygen atom (H---O: 1.879 Å); and (ii) contacts between 3’-NH2 and O4 atom of T8 (H---O: 2.849 Å) [18].
One of the earliest computational studies on platinated-DNA (cisplatin and oxaliplatin), using the same 12-mer DNA sequence as used in this study and in crystal and NMR studies, was carried out by Howell’s group [19]. Based on their study on cisplatin-DNA adduct they reported the following: (a) no hydrogen-bond contacts between 5’-NH3 hydrogens of cisplatin and g6 phosphate backbone oxygen atoms, although these interactions were seen in the x-ray structure of cisplatin-DNA; (b) hydrogen-bond contacts between 3’-NH3 hydrogens and O6 atom of g7; (c) the modeled structure had normal Watson-Crick base-pairing between T8-A17, unlike that found in the crystal structure of cisplatin-DNA. Also, these authors reported the following computational results on oxaliplatin-DNA adduct: (a) no hydrogen-bond contacts between 5’-NH2 hydrogens and g6 phosphate backbone oxygen atoms; and (b) hydrogen-bond contacts between 3’- NH2 hydrogens of DACH and O4 atom of T8.
In a recent study, Chaney et al. reported combined NMR and MD data for cisplatin-DNA and oxaliplatin-DNA (using the same sequence as used in this study) [21]. From the MD studies, no hydrogen-bond contacts were identified between platinum ligands and the 5’-DNA bases; whereas hydrogen-bond contacts between platinum ligands and the 3’-DNA bases were identified. These authors also explained that the lack of preferential binding of high mobility group box proteins (HMGB1a) to cisplatin-TGGT and oxaliplatin-TGGT was due to the increased conformational flexibility caused by the hydrogen-bonds between platinum ligands and 3’-DNA bases. Using pyrazolatoplatinum- DNA adducts, Komeda et al. showed that when reacted with DNA, the pyrazolato-platinum derivatives did not deform DNA as was shown with cisplatin or oxaliplatin. Additionally, these pyrazolato platinum compounds exhibited good cytotoxicity activity [22]. Since we knew that BNP3029 was more potent in comparison to cisplatin and oxaliplatin based on cytotoxicity studies in a variety of wild-type and resistant human cancer cell lines [6], we thought it would be of interest to study the platination of DNA by BNP3029 in comparison to the platination of DNA by cisplatin or oxaliplatin.
Here, we present data describing how platinum-DNA adducts involving cisplatin, oxaliplatin, and BNP3029 affect DNA morphology in relation to normal B-DNA. Several molecular dynamics simulations (either 60 ns or 10 ns) were carried out with 5’-CCTCTggTCTCC-3’ (where ’gg’ represents any of the 3 platinum-DNA adducts: gg- Pt(NH3)2 for cisplatin; gg-Pt(DACH) for oxaliplatin; and gg- Pt[N≡C(CH2)3(C6H5)]2 for BNP3029) using electrostatic potential (ESP) or restrained electrostatic potential (RESP) charges. As our goal was to compare the morphological changes in platinated-DNA with B-DNA, simulations using a B-form DNA with the same sequence were also conducted. Since DNA is known to have fluxional behavior [23-27], detailed statistical analyses were carried out using the Kolmogorov- Smirnov (KS) scheme to compare the morphological parameters from platinated-DNA MD runs with B-DNA MD runs.
Even though there are 42 morphological parameters for DNA to monitor the changes leading to B-DNA or A-DNA, analyses herein focused on the MD runs on those parameters that highlight the differences between a B-form or an A-form DNA such as changes in: inclination (η), twist (Ω), roll (ρ), slide (Dy), rise (Dz); major and minor groove widths and depths; backbone angles delta (δ), zeta (ζ), and chi (χ); and sugar pucker upon platination. Initially, the following were presented from the MD simulations on platinated-DNA: (i) root-mean-square deviations; (ii) Watson-Crick base-pairing; (iii) hydrogen-bond contacts between platinum ligand atoms and DNA; (iv) The ligand arrangement specific to BNP3029; and (v) sequenceaveraged morphological data. Later, the morphological changes in individual bases from the MD runs were highlighted. Unless specified, all the results presented herein for the platinated-DNA adducts were based on MD runs obtained using the RESP charges. For B-DNA, the parm99 forcefield was used.
RMSD for B-DNA, cisplatin-DNA, oxaliplatin-DNA and BNP3029-DNA
The root-mean-square deviation (RMSD) plots provide information about how well the system equilibrated and for the presence of any substates during the simulation time. The RMSDs of B-DNA or platinated-DNA (from the inital structure using all the atoms for RMSD and using the 60 ns MD runs) is in the range of 4-6 Å (Figure 2). This RMSD is similar to the RMSD obtained on B-DNA in an earlier study when the parmbsc0 forcefield was included in the MD runs [27]. The RMSD plots of cisplatin-DNA and oxaliplatin- DNA averaged around 5 Å and did not show any significant substates whereas the RMSD plot of BNP3029-DNA did indicate substates during the course of the simulation (near 20 ns and 40 ns) possibly due to the positioning of the substituted cyano ligand of BNP3029 near DNA bases and backbone. A superimposition of the structures after every 1 ns revealed an interesting detail. The superimposed structures from BNP3029-DNA showed that BNP3029 carrier ligands (two substituted cyano groups) occupy the entire major groove area near g6g7 compared to the limited space occupied by the carrier ligands of cisplatin (two NH3s) and oxaliplatin (1,2-diaminocyclohexane, DACH) (Figure 3). The occupation of the major groove near gg by the substituted cyano ligands of BNP3029 may prevent the preferential binding of subsequent cellular machinery to DNA and which in turn may provide enhanced cytotoxic activity for BNP3029.
Figure 2: 2A Root-mean-square deviation (RMSD in Å) for B-DNA using the 60 ns MD run. 2B Root-mean-square deviation (RMSD in Å) for cisplatin-DNA using the 60 ns MD run with RESP charges. 2C Root-mean-square deviation (RMSD in Å) for oxaliplatin-DNA using the 60 ns MD run with RESP charges. 2D Rootmean- square deviation (RMSD in Å) for BNP3029-DNA using the 60 ns MD run with RESP charges
Percentage (%) occupancy of hydrogen-bonds between base pairs
The percentage occupation of hydrogen-bonds (obtained using all the uncorrelated samples from RESP MD runs) between various basepairs did not indicate any differences in the hydrogen-bond occupancy on the 5’-side of the platinated-DNA compared to the similar occupancy on the 5’-side of B-DNA (Figure 4, panel A). Of the three possible hydrogen-bonds between the G-C base-pair, the g6 base in platinated- DNA from cisplatin, oxaliplatin and BNP3029 showed a slight drop in the occupancy of the hydrogen-bond between N2(H21) atom of g6 and O2 atom of C19 compared to the one in B-DNA. The other two hydrogen-bonds in platinated-DNA did not show any significant occupancy change from B-DNA for this base pair. The hydrogenbond between O4 atom of T8 and N6(H61) atom of A17 also showed lower occupancy in the platinated-DNA from cisplatin and oxaliplatin compared to either B-DNA or platinated-DNA from BNP3029. A plausible reason for the lower occupancy for this hydrogen-bond in oxaliplatin-DNA or cisplatin-DNA was due to the enhanced hydrogenbonding between the O4 atom of T8 with one of the NH2 hydrogens on DACH in oxaliplatin or with the NH3 hydrogens of cisplatin. Because of this interaction, the base-pair loses some of the hydrogen-bonding contributions between O4 atom of T8 and N6(H61) atom of A17. As noted previously for the hydrogen-bond occupancy, there is not much of a change in the hydrogen-bond distance patterns between base-pairs in platinated-DNA compared to normal B-DNA (Figure 4, panel B).
Figure 4: Averaged hydrogen-bonds between Watson-Crick base-pairs (panel A fraction occupied and panel B average hydrogen-bond distance in Å) for the 12- mer DNA (5’-CCTCTggTCTCC-3’) from the platinated-DNA and B-DNA. The average data were generated using the uncorrelated samples from five MD runs using the RESP charges for the platinated-DNA. Panels A and B contain data for Bases 2 to 11 only.
Hydrogen-bonds between bases of DNA and the carrier ligands of cisplatin and oxaliplatin
The current MD studies on platinated-DNA for cisplatin and oxaliplatin did not indicate significant hydrogen-bond patterns on the 5’-side of the platinum adduct (similar to the earlier results presented by Howell [19] and Chaney [21]). However, as shown in Figure 2S, we identified two different hydrogen-bonding patterns on the 3’-side of the platinum-DNA with cisplatin and oxaliplatin. Oxaliplatin has a slightly higher frequency for both the hydrogen-bonds. The first one is between the O6 atom of g7 base and the NH2 hydrogens of DACH (or all three NH3 hydrogens in the case of cisplatin) and the second one is between the O4 atom of T8 base and the NH2 hydrogens of DACH (or all three NH3 hydrogens in the case of cisplatin). For the hydrogenbond between the O6 atom of g7 base and the NH2 hydrogens, oxaliplatin has an occupancy (obtained using a single 60 ns MD run) of 62.9% vs. 48.1% (for cisplatin, all three NH3 hydrogens combined). In the case of the second hydrogen-bond between the O4 atom of T8 base and the NH2 hydrogens, the occupancies are 60.8% and 56.6% for oxaliplatin and cisplatin-DNA respectively (for cisplatin, all three NH3 hydrogens were combined).
C-H---O elecrostatic interactions between DNA and the carrier ligands of BNP3029
Since the substituted cyano group (carrier ligand of BNP3029) did not have any hydrogens that could potentially interact with the O6 atom of g7 base or O4 atom of T8 base, other stabilizing interactions are monitored such as C-H---O bonds. One of the three CH2s or the phenyl group in BNP3029 could potentially provide the C-H bond and the O atom could come from either a base or from the backbone of DNA. No significant populations of C-H (from BNP3029) ---O (DNA bases/backbone) type electrostatic interactions, that would indicate any stabilizing interactions between the side chains of BNP3029 and DNA bases, were noted.
π-π stacking between the phenyl rings of BNP3029
π-π interactions involving aromatic ring systems are very important in several chemistry reactions [28]. Based upon theoretical and experimental studies on benzene dimers [28] for a description of various benzene dimer configurations], it has been shown that the T-shaped arrangement of two benzene rings is the global energy minimum (in T-shaped arrangement one C-H hydrogen from one benzene ring is directed towards the other benzene ring). Also the Y-shaped arrangement, in which two hydrogens from one benzene ring are directed towards the other benzene ring resembling a Y-shape, and the parallel-displaced configuration of two benzene rings are both close to the global energy minimum. Sampling the conformational space occupied by the two phenyl rings of BNP3029-DNA MD runs (combining all the uncorrelated samples) indicated that approximately 40% of the time the two phenyl rings are either in T-, Y-, or in paralleldisplaced configuration.
Changes in DNA morphology upon platination
The sequence-averaged morphological parameters for B-DNA and platinated-DNA are presented in Tables 1-4 (data for the bases T3 to T10 are averaged, excluding the two end bases on either side of the platinum adduct, for a total of eight bases). For each of the five MD runs for B-DNA or platinated-DNA adducts with cisplatin, oxaliplatin and BNP3029, sets of uncorrelated samples were obtained and combined in generating the sequence-averaged data. Table 1 compares the sequence-averaged morphological data for the B-DNA MD run with the corresponding data for DNA obtained from NMR data (same sequence, pdb code: 2K0V). As seen in Table 1, most of the parameters for B-DNA from NMR (2K0V) data are within one standard deviation (SD) from the mean values obtained using the MD runs on B-DNA. Since the inclusion of parmbsc0 forcefield in the MD runs maintains alphaW and alphaC, and gammaW and gammaC backbone angles around g- (-60)/g+ (+60) values respectively, the sequence-averaged MD data for these parameters are closer to the above mentioned values compared to the values from the NMR data.
| 2K0V | Morphological Parameter | Mean | SD | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|
| 11.10 | tbend | 16.61 | 10.70 | 0.60 | 8.72 | 14.28 | 21.53 | 52.55 |
| -0.03 | shear | -0.06 | 0.30 | -1.01 | -0.25 | -0.06 | 0.15 | 0.86 |
| -0.21 | stretch | 0.02 | 0.12 | -0.40 | -0.05 | 0.01 | 0.09 | 0.89 |
| 0.00 | stagger | 0.08 | 0.40 | -1.25 | -0.18 | 0.08 | 0.34 | 1.54 |
| -0.10 | buckle | -3.08 | 11.48 | -35.21 | -10.85 | -3.60 | 4.90 | 28.47 |
| -0.60 | propel | -11.52 | 8.97 | -40.00 | -17.72 | -11.89 | -5.63 | 23.00 |
| -1.00 | opening | 2.18 | 4.90 | -14.89 | -0.82 | 1.87 | 4.96 | 27.39 |
| -0.77 | xdisp | -1.96 | 0.96 | -4.86 | -2.65 | -1.98 | -1.33 | 1.59 |
| -0.07 | ydisp | 0.06 | 0.66 | -2.30 | -0.38 | 0.06 | 0.49 | 2.10 |
| 4.60 | inclin | 8.30 | 6.42 | -7.80 | 4.35 | 8.06 | 11.62 | 45.31 |
| -3.80 | tip | -0.93 | 5.27 | -36.27 | -4.24 | -0.85 | 2.26 | 15.16 |
| 2.50 | ax-bend | 1.97 | 1.34 | 0.02 | 1.06 | 1.69 | 2.53 | 9.30 |
| 0.08 | shift | 0.08 | 0.74 | -2.03 | -0.43 | 0.09 | 0.62 | 2.90 |
| -0.21 | slide | -0.60 | 0.80 | -2.85 | -1.16 | -0.62 | -0.09 | 3.19 |
| 3.21 | rise | 3.33 | 0.34 | 2.35 | 3.11 | 3.33 | 3.53 | 4.52 |
| 0.40 | tilt | 0.67 | 4.48 | -13.24 | -2.42 | 0.29 | 3.65 | 18.52 |
| 1.40 | roll | 3.78 | 6.20 | -16.47 | -0.60 | 3.52 | 8.07 | 26.89 |
| 34.70 | twist | 31.07 | 8.84 | -33.62 | 28.03 | 31.68 | 35.76 | 49.85 |
| 3.22 | h-ris | 3.23 | 0.36 | 1.99 | 2.98 | 3.23 | 3.45 | 4.77 |
| 35.00 | h-twi | 31.50 | 8.66 | -32.17 | 28.45 | 32.15 | 36.30 | 49.91 |
| -47.30 | alphaW | -69.65 | 12.23 | -167.45 | -76.33 | -68.44 | -61.33 | -34.08 |
| -179.60 | betaW | 171.05 | 12.69 | 53.74 | 164.44 | 171.28 | 178.50 | 207.66 |
| 38.10 | gammaW | 57.51 | 11.71 | 10.68 | 50.80 | 57.72 | 63.37 | 191.00 |
| 134.20 | deltaW | 114.07 | 20.81 | 63.89 | 95.80 | 116.27 | 130.99 | 169.16 |
| 172.40 | epsilW | -170.11 | 20.55 | -207.39 | -180.02 | -173.09 | -166.41 | -53.01 |
| -98.50 | zetaW | -94.04 | 31.77 | -305.19 | -96.18 | -88.00 | -80.56 | -41.65 |
| -111.00 | chiW | -126.29 | 19.21 | -183.39 | -140.53 | -127.10 | -114.39 | -44.55 |
| 154.20 | phaseW | 118.78 | 38.89 | 1.79 | 93.34 | 122.26 | 145.49 | 357.19 |
| 32.20 | ampW | 38.50 | 6.83 | 15.42 | 34.06 | 38.64 | 43.52 | 55.94 |
| -38.30 | alphaC | -72.38 | 16.15 | -309.42 | -78.97 | -70.29 | -62.95 | -37.29 |
| -175.60 | betaC | 170.00 | 16.13 | 58.83 | 163.47 | 170.81 | 179.18 | 222.30 |
| 30.90 | gammaC | 56.75 | 16.09 | 12.16 | 49.41 | 56.25 | 62.74 | 291.07 |
| 142.60 | deltaC | 120.27 | 21.59 | 63.57 | 103.19 | 124.86 | 138.67 | 161.37 |
| 175.50 | epsilC | -164.88 | 27.56 | -205.05 | -179.32 | -172.25 | -164.15 | -59.33 |
| -105.80 | zetaC | -101.85 | 39.55 | -279.14 | -101.16 | -89.97 | -81.97 | -50.18 |
| -107.00 | chiC | -120.02 | 22.11 | -174.82 | -136.00 | -119.26 | -105.49 | -21.79 |
| 168.20 | phaseC | 129.77 | 38.66 | 4.69 | 101.85 | 133.88 | 155.72 | 307.24 |
| 34.80 | ampC | 38.67 | 7.18 | 12.01 | 33.85 | 39.12 | 43.88 | 56.39 |
| 7.41 | minw | 6.97 | 1.65 | 0.00 | 5.90 | 7.18 | 8.05 | 11.60 |
| 4.52 | mind | 4.16 | 0.91 | 0.00 | 3.67 | 4.24 | 4.72 | 6.64 |
| 10.22 | majw | 10.06 | 5.55 | 0.00 | 9.98 | 12.13 | 13.56 | 19.27 |
| 5.08 | majd | 4.58 | 3.32 | -1.50 | 1.54 | 5.31 | 7.45 | 10.89 |
Table 1: Sequence-averaged morphological parameters for B-DNA using the uncorrelated snap shots from five MD simulations (total length of MD run is 100 ns). 2K0V numbers refer to the NMR data. The 2K0V numbers highlighted in green are within Mean ± SD values from the MD run. SD is the standard deviation. Min and Max are the minimum and maximum values obtained for that parameter from the MD run. 25%, Median (50%) and 75% are the respective percentiles between Min and Max. tbend is total bend. Ax-bend is a re-rendering of the total bend using the local base-step data. Helical-Rise and Helical-Twist correspond to the translation and rotation of successive base pairs along and around the helical axis. Backbone angles are presented separately for “W” (platinated strand) and “C” (complementary strand). Under Morphological Parameter column, min and maj refer to minor and major grooves and w and d stand for width and depth.
| 2K0V | 3LPV_molA | 3LPV_molB | 1A84 | Morphological Parameter | Mean | SD | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11.10 | 29.50 | 27.60 | 83.20 | tbend | 32.91 | 13.47 | 1.64 | 23.60 | 32.74 | 42.30 | 83.52 |
| -0.03 | -0.05 | -0.10 | 0.03 | shear | -0.09 | 0.31 | -1.84 | -0.30 | -0.08 | 0.11 | 0.93 |
| -0.21 | -0.12 | -0.13 | -0.17 | stretch | 0.02 | 0.13 | -0.37 | -0.06 | 0.01 | 0.09 | 0.98 |
| 0.00 | 0.13 | 0.10 | -0.20 | stagger | 0.10 | 0.45 | -3.16 | -0.19 | 0.10 | 0.40 | 1.81 |
| -0.10 | -1.30 | 0.00 | 6.90 | buckle | -5.23 | 13.76 | -55.52 | -15.09 | -5.21 | 4.75 | 37.53 |
| -0.60 | -13.00 | -12.60 | -2.00 | propel | -11.01 | 11.02 | -51.94 | -18.50 | -11.42 | -3.66 | 25.61 |
| -1.00 | 2.50 | 2.60 | 4.00 | opening | 2.80 | 5.35 | -25.28 | -0.49 | 2.39 | 5.67 | 45.44 |
| -0.77 | -2.98 | -3.07 | -2.11 | xdisp | -1.95 | 0.93 | -4.88 | -2.56 | -1.93 | -1.33 | 1.38 |
| -0.07 | 0.64 | 0.62 | 0.38 | ydisp | -0.01 | 0.80 | -2.77 | -0.59 | -0.02 | 0.58 | 2.42 |
| 4.60 | 14.20 | 14.80 | 20.50 | inclin | 13.68 | 6.98 | -9.92 | 9.05 | 13.75 | 18.49 | 36.11 |
| -3.80 | -2.20 | -2.20 | -2.90 | tip | -2.51 | 9.64 | -30.86 | -8.87 | -2.40 | 3.97 | 27.22 |
| 2.50 | 3.30 | 3.10 | 10.70 | ax-bend | 4.27 | 1.85 | 0.06 | 2.96 | 4.10 | 5.36 | 12.15 |
| 0.08 | -0.03 | 0.03 | -0.06 | shift | 0.23 | 0.84 | -2.69 | -0.36 | 0.22 | 0.82 | 3.85 |
| -0.21 | -1.03 | -1.03 | -0.20 | slide | -0.51 | 0.74 | -2.51 | -1.03 | -0.55 | -0.07 | 3.60 |
| 3.21 | 3.23 | 3.24 | 3.83 | rise | 3.40 | 0.52 | 2.20 | 3.07 | 3.32 | 3.64 | 5.72 |
| 0.40 | 1.50 | 1.50 | 0.90 | tilt | 1.36 | 4.91 | -25.19 | -1.88 | 1.34 | 4.53 | 23.51 |
| 1.40 | 7.80 | 8.00 | 5.40 | roll | 7.36 | 14.46 | -21.91 | -0.83 | 3.83 | 9.18 | 59.71 |
| 34.70 | 32.10 | 32.20 | 28.10 | twist | 30.20 | 7.63 | -28.38 | 25.53 | 30.31 | 35.47 | 58.53 |
| 3.22 | 2.77 | 2.76 | 3.34 | h-ris | 3.18 | 0.52 | 1.61 | 2.83 | 3.17 | 3.50 | 5.32 |
| 35.00 | 33.60 | 33.60 | 32.30 | h-twi | 31.64 | 6.74 | -30.58 | 27.59 | 31.61 | 35.90 | 57.16 |
| -47.30 | -71.10 | -68.90 | -66.40 | alphaW | -71.96 | 18.04 | -307.39 | -77.88 | -70.06 | -62.95 | -36.93 |
| -179.60 | 177.50 | 177.70 | 162.80 | betaW | 169.86 | 18.09 | 41.26 | 163.75 | 171.14 | 178.22 | 300.13 |
| 38.10 | 62.50 | 61.60 | 113.50 | gammaW | 59.39 | 18.01 | 4.55 | 51.39 | 57.50 | 64.00 | 201.67 |
| 134.20 | 90.20 | 91.60 | 132.40 | deltaW | 109.96 | 21.79 | 62.37 | 91.45 | 109.68 | 128.68 | 163.65 |
| 172.40 | -158.20 | -157.70 | -177.90 | epsilW | -167.65 | 21.72 | -336.13 | -178.51 | -171.41 | -163.38 | -60.10 |
| -98.50 | -75.90 | -77.00 | -98.90 | zetaW | -90.76 | 29.88 | -299.73 | -95.24 | -86.40 | -76.81 | -43.63 |
| -111.00 | -155.00 | -153.40 | -111.30 | chiW | -127.62 | 19.04 | -176.07 | -141.38 | -130.58 | -115.29 | -53.96 |
| 154.20 | 21.70 | 17.70 | 168.90 | phaseW | 107.97 | 45.32 | 0.60 | 80.11 | 114.86 | 139.56 | 358.54 |
| 32.20 | 40.40 | 40.70 | 26.20 | ampW | 39.56 | 6.87 | 5.68 | 35.24 | 39.97 | 44.49 | 59.51 |
| -38.30 | -80.20 | -71.80 | -90.20 | alphaC | -75.26 | 15.66 | -285.51 | -83.00 | -73.04 | -65.41 | -33.86 |
| -175.60 | 171.10 | 173.90 | -174.70 | betaC | 170.69 | 14.01 | 70.34 | 163.03 | 171.27 | 179.12 | 239.61 |
| 30.90 | 62.40 | 57.90 | 98.50 | gammaC | 55.04 | 11.56 | 3.05 | 48.32 | 54.72 | 61.63 | 193.47 |
| 142.60 | 94.10 | 94.00 | 136.00 | deltaC | 118.50 | 23.13 | 64.03 | 97.10 | 124.19 | 138.11 | 160.58 |
| 175.50 | -150.70 | -157.10 | 171.40 | epsilC | -160.55 | 29.65 | -203.66 | -176.73 | -169.20 | -159.50 | -50.24 |
| -105.80 | -83.40 | -83.30 | -80.00 | zetaC | -102.51 | 42.91 | -284.62 | -100.57 | -88.77 | -79.50 | -42.76 |
| -107.00 | -148.70 | -147.40 | -108.20 | chiC | -118.79 | 20.35 | -168.95 | -134.64 | -118.84 | -104.22 | -17.41 |
| 168.20 | 34.10 | 34.10 | 162.70 | phaseC | 123.93 | 46.76 | 1.41 | 93.45 | 133.89 | 155.81 | 359.86 |
| 34.80 | 45.60 | 42.90 | 27.70 | ampC | 37.84 | 7.45 | 2.22 | 33.20 | 38.16 | 42.91 | 57.58 |
| 7.41 | 9.48 | 9.60 | 7.99 | minw | 8.12 | 1.98 | 0.00 | 6.77 | 8.10 | 9.65 | 12.88 |
| 4.52 | 1.83 | 1.72 | 2.73 | mind | 3.56 | 1.19 | -0.15 | 2.85 | 3.77 | 4.40 | 7.09 |
| 10.22 | 4.94 | 5.23 | 10.12 | majw | 9.69 | 5.08 | 0.00 | 7.91 | 11.42 | 13.34 | 19.78 |
| 5.08 | 9.67 | 9.71 | 11.88 | majd | 5.22 | 3.99 | -0.94 | 1.46 | 4.85 | 8.90 | 14.05 |
Table 2: Sequence-averaged morphological parameters for Cisplatin-DNA using the uncorrelated snap shots from five MD simulations (total length of MD run is 150 ns, RESP charges were used for (NH3)2Ptgg). 2K0V is unplatinated DNA from NMR studies.1A84 numbers refer to the NMR data. 3LPV numbers refer to the X-ray data. The 2K0V, 3LPV and 1A84 numbers highlighted in green are within Mean ± SD values from the MD run. SD is the standard deviation. Min and Max are the minimum and maximum values obtained for that parameter from the MD run. 25%, Median (50%) and 75% are the respective percentiles between Min and Max. tbend is total bend. Ax-bend is a re-rendering of the total bend using the local base-step data. Helical-Rise and Helical-Twist correspond to the translation and rotation of successive base pairs along and around the helical axis. Backbone angles are presented separately for “W” (platinated strand) and “C” (complementary strand). Under Morphological Parameter column, min and maj refer to minor and major grooves and w and d stand for width and depth.
| 2K0V | 1IHH | 2K0U | Morphological Parameter | Mean | SD | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|---|---|
| 11.10 | 24.50 | 35.60 | tbend | 31.21 | 12.41 | 0.81 | 22.81 | 30.89 | 39.47 | 75.12 |
| -0.03 | 0.06 | -0.04 | shear | -0.09 | 0.31 | -2.30 | -0.29 | -0.08 | 0.11 | 1.27 |
| -0.21 | -0.22 | -0.22 | stretch | 0.02 | 0.13 | -0.48 | -0.07 | 0.01 | 0.09 | 1.39 |
| 0.00 | 0.29 | 0.02 | stagger | 0.14 | 0.47 | -2.24 | -0.16 | 0.14 | 0.45 | 1.86 |
| -0.10 | -4.50 | -0.60 | buckle | -6.30 | 13.61 | -55.75 | -15.94 | -6.31 | 3.38 | 35.48 |
| -0.60 | -9.60 | -0.80 | propel | -10.82 | 10.73 | -51.73 | -18.49 | -11.26 | -3.55 | 32.15 |
| -1.00 | 0.10 | 0.20 | opening | 2.61 | 5.32 | -37.54 | -0.79 | 2.17 | 5.56 | 36.75 |
| -0.77 | -2.94 | -1.71 | xdisp | -1.96 | 0.92 | -5.01 | -2.58 | -1.95 | -1.34 | 1.33 |
| -0.07 | 0.60 | 0.03 | ydisp | -0.12 | 0.78 | -3.02 | -0.67 | -0.13 | 0.43 | 2.75 |
| 4.60 | 11.10 | 20.00 | inclin | 13.30 | 6.68 | -22.23 | 9.09 | 13.85 | 17.92 | 36.91 |
| -3.80 | -3.60 | 5.00 | tip | -2.48 | 9.99 | -30.93 | -8.90 | -2.46 | 4.25 | 30.27 |
| 2.50 | 2.70 | 2.90 | ax-bend | 4.23 | 1.72 | 0.19 | 3.03 | 4.07 | 5.24 | 11.52 |
| 0.08 | 0.05 | -0.22 | shift | 0.21 | 0.79 | -2.22 | -0.34 | 0.21 | 0.80 | 2.66 |
| -0.21 | -1.11 | 0.31 | slide | -0.57 | 0.66 | -2.43 | -1.05 | -0.59 | -0.12 | 2.82 |
| 3.21 | 3.22 | 3.77 | rise | 3.41 | 0.53 | 2.11 | 3.07 | 3.33 | 3.64 | 5.98 |
| 0.40 | 2.30 | 0.70 | tilt | 1.24 | 4.93 | -17.35 | -2.15 | 1.12 | 4.61 | 23.60 |
| 1.40 | 5.10 | 11.10 | roll | 7.10 | 15.12 | -22.52 | -1.17 | 3.15 | 8.27 | 65.56 |
| 34.70 | 32.10 | 31.20 | twist | 30.40 | 6.85 | -9.62 | 25.78 | 30.40 | 35.41 | 50.79 |
| 3.22 | 2.80 | 3.56 | h-ris | 3.19 | 0.54 | 1.41 | 2.83 | 3.18 | 3.52 | 5.71 |
| 35.00 | 33.00 | 32.60 | h-twi | 31.75 | 5.78 | -2.04 | 28.01 | 31.52 | 35.69 | 49.26 |
| -47.30 | -74.60 | -57.60 | alphaW | -70.59 | 12.94 | -291.02 | -76.81 | -69.79 | -62.97 | -34.09 |
| -179.60 | 165.70 | 170.60 | betaW | 170.41 | 12.62 | 52.92 | 163.70 | 170.57 | 177.91 | 225.60 |
| 38.10 | 70.10 | 50.50 | gammaW | 58.14 | 10.75 | 17.99 | 52.07 | 58.04 | 63.92 | 184.72 |
| 134.20 | 85.90 | 121.60 | deltaW | 108.93 | 21.62 | 59.38 | 90.13 | 109.33 | 127.07 | 163.16 |
| 172.40 | -159.80 | -159.80 | epsilW | -168.08 | 19.24 | -211.98 | -178.08 | -171.46 | -164.20 | -55.49 |
| -98.50 | -70.20 | -100.90 | zetaW | -90.22 | 27.11 | -257.03 | -95.66 | -86.51 | -76.96 | -38.12 |
| -111.00 | -158.90 | -122.20 | chiW | -129.49 | 17.12 | -178.11 | -142.20 | -131.23 | -118.48 | -62.44 |
| 154.20 | 34.50 | 131.70 | phaseW | 105.17 | 45.33 | 0.40 | 74.18 | 113.53 | 137.96 | 359.68 |
| 32.20 | 35.40 | 31.00 | ampW | 39.14 | 6.77 | 5.17 | 34.76 | 39.47 | 43.92 | 59.40 |
| -38.30 | -65.40 | -54.60 | alphaC | -75.06 | 15.98 | -284.28 | -81.74 | -73.10 | -65.39 | -32.77 |
| -175.60 | 170.40 | 161.80 | betaC | 171.04 | 13.98 | 50.17 | 163.61 | 171.38 | 179.35 | 241.31 |
| 30.90 | 49.40 | 47.40 | gammaC | 55.10 | 12.79 | 5.89 | 48.30 | 55.17 | 61.58 | 358.08 |
| 142.60 | 93.60 | 99.00 | deltaC | 117.76 | 23.48 | 62.11 | 95.84 | 123.29 | 137.94 | 164.84 |
| 175.50 | -158.80 | -155.50 | epsilC | -161.05 | 28.88 | -208.20 | -176.57 | -169.07 | -160.25 | -33.79 |
| -105.80 | -79.00 | -93.60 | zetaC | -100.76 | 41.72 | -274.05 | -99.02 | -87.55 | -78.91 | -46.58 |
| -107.00 | -146.60 | -131.80 | chiC | -119.38 | 19.97 | -173.76 | -134.49 | -119.97 | -105.07 | -45.90 |
| 168.20 | 43.20 | 48.30 | phaseC | 122.95 | 48.67 | 0.27 | 88.51 | 132.90 | 155.86 | 359.77 |
| 34.80 | 40.70 | 32.90 | ampC | 37.61 | 7.37 | 7.43 | 32.92 | 37.88 | 42.75 | 59.09 |
| 7.41 | 9.52 | 11.70 | minw | 8.27 | 1.95 | 0.00 | 6.92 | 8.28 | 9.78 | 13.23 |
| 4.52 | 2.15 | 1.08 | mind | 3.53 | 1.19 | -2.73 | 2.78 | 3.78 | 4.42 | 6.63 |
| 10.22 | 8.02 | 9.76 | majw | 11.09 | 4.01 | 0.00 | 9.87 | 11.81 | 13.72 | 17.42 |
| 5.08 | 7.96 | 4.88 | majd | 5.29 | 4.00 | -2.08 | 1.05 | 5.42 | 8.94 | 13.50 |
Table 3: Sequence-averaged morphological parameters for Oxaliplatin-DNA using the uncorrelated snap shots from five MD simulations (total length of MD run is 150 ns, RESP charges were used for (DACH)Ptgg). 2K0V is unplatinated DNA from NMR studies.2K0U numbers refer to the NMR data. 1IHH numbers refer to the X-ray data. The 2K0V, 2K0U and 1IHH numbers highlighted in green are within Mean ± SD values from the MD run. SD is the standard deviation. Min and Max are the minimum and maximum values obtained for that parameter from the MD run. 25%, Median (50%) and 75% are the respective percentiles between Min and Max. tbend is total bend. Axbend is a re-rendering of the total bend using the local base-step data. Helical-Rise and Helical-Twist correspond to the translation and rotation of successive base pairs along and around the helical axis. Backbone angles are presented separately for “W” (platinated strand) and “C” (complementary strand). Under Morphological Parameter column, min and maj refer to minor and major grooves and w and d stand for width and depth.
| 2K0V | Morphological Parameter | Mean | SD | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|
| 11.1 | tbend | 28.22 | 12.25 | 1.25 | 19.83 | 26.66 | 35.37 | 62.30 |
| -0.03 | shear | -0.10 | 0.32 | -2.87 | -0.30 | -0.09 | 0.10 | 0.82 |
| -0.21 | stretch | 0.02 | 0.13 | -0.42 | -0.06 | 0.01 | 0.09 | 1.55 |
| 0 | stagger | 0.10 | 0.49 | -2.30 | -0.22 | 0.10 | 0.42 | 2.11 |
| -0.1 | buckle | -4.91 | 13.92 | -56.96 | -14.85 | -5.77 | 4.83 | 39.97 |
| -0.6 | propel | -11.11 | 11.20 | -49.09 | -18.49 | -11.10 | -3.77 | 36.15 |
| -1 | opening | 2.50 | 5.29 | -29.70 | -0.58 | 2.31 | 5.37 | 49.42 |
| -0.77 | xdisp | -2.10 | 0.90 | -5.13 | -2.73 | -2.10 | -1.49 | 0.95 |
| -0.07 | ydisp | -0.09 | 0.80 | -2.91 | -0.65 | -0.07 | 0.49 | 2.93 |
| 4.6 | inclin | 12.10 | 7.08 | -8.18 | 7.66 | 12.26 | 16.76 | 36.17 |
| -3.8 | tip | -3.41 | 8.02 | -32.63 | -8.89 | -3.34 | 2.14 | 20.80 |
| 2.5 | ax-bend | 3.62 | 1.66 | 0.09 | 2.39 | 3.44 | 4.64 | 10.74 |
| 0.08 | shift | 0.25 | 0.84 | -2.13 | -0.34 | 0.21 | 0.86 | 3.10 |
| -0.21 | slide | -0.61 | 0.77 | -2.56 | -1.20 | -0.60 | -0.06 | 2.22 |
| 3.21 | rise | 3.36 | 0.46 | 2.21 | 3.05 | 3.33 | 3.62 | 6.12 |
| 0.4 | tilt | 1.56 | 5.24 | -20.36 | -1.80 | 1.43 | 4.77 | 20.19 |
| 1.4 | roll | 6.17 | 12.13 | -34.79 | -1.22 | 3.38 | 9.58 | 60.48 |
| 34.7 | twist | 30.17 | 7.25 | -4.58 | 25.85 | 30.20 | 35.26 | 49.93 |
| 3.22 | h-ris | 3.19 | 0.46 | 1.86 | 2.90 | 3.17 | 3.46 | 5.66 |
| 35 | h-twi | 31.40 | 6.48 | 0.32 | 27.40 | 31.54 | 35.76 | 50.79 |
| -47.3 | alphaW | -68.96 | 11.35 | -149.03 | -75.35 | -68.49 | -61.74 | -36.30 |
| -179.6 | betaW | 170.70 | 12.50 | 105.54 | 163.64 | 171.56 | 178.70 | 222.40 |
| 38.1 | gammaW | 57.12 | 9.57 | 3.85 | 51.24 | 57.21 | 63.20 | 92.32 |
| 134.2 | deltaW | 111.83 | 20.69 | 66.88 | 94.29 | 111.53 | 129.48 | 156.83 |
| 172.4 | epsilW | -170.51 | 19.07 | -205.11 | -180.17 | -173.05 | -165.77 | -56.52 |
| -98.5 | zetaW | -92.14 | 25.74 | -237.66 | -96.69 | -88.64 | -80.39 | -39.15 |
| -111 | chiW | -124.85 | 20.29 | -175.40 | -140.31 | -127.71 | -112.33 | -60.53 |
| 154.2 | phaseW | 114.74 | 35.08 | 2.08 | 91.44 | 117.49 | 140.82 | 338.96 |
| 32.2 | ampW | 40.13 | 6.92 | 14.25 | 35.78 | 40.60 | 44.97 | 59.19 |
| -38.3 | alphaC | -76.94 | 23.59 | -287.85 | -83.64 | -73.87 | -65.29 | -29.21 |
| -175.6 | betaC | 171.05 | 14.75 | 73.47 | 163.20 | 171.02 | 180.11 | 248.48 |
| 30.9 | gammaC | 56.41 | 20.90 | 3.28 | 47.97 | 55.23 | 61.78 | 359.39 |
| 142.6 | deltaC | 120.72 | 21.78 | 61.40 | 102.44 | 126.74 | 138.38 | 166.33 |
| 175.5 | epsilC | -159.55 | 31.40 | -207.36 | -177.09 | -169.63 | -158.81 | -50.12 |
| -105.8 | zetaC | -104.87 | 44.79 | -273.89 | -101.98 | -89.42 | -81.07 | -48.78 |
| -107 | chiC | -118.44 | 21.32 | -172.54 | -135.01 | -119.13 | -103.21 | -36.69 |
| 168.2 | phaseC | 129.24 | 42.13 | 2.38 | 102.72 | 136.17 | 156.56 | 355.44 |
| 34.8 | ampC | 38.30 | 7.56 | 1.97 | 33.38 | 38.49 | 43.75 | 58.20 |
| 7.41 | minw | 7.63 | 1.87 | 0.00 | 6.37 | 7.69 | 8.81 | 12.47 |
| 4.52 | mind | 3.82 | 1.08 | -2.86 | 3.21 | 3.96 | 4.56 | 7.27 |
| 10.22 | majw | 10.50 | 5.06 | 0.00 | 9.55 | 11.69 | 13.82 | 19.94 |
| 5.08 | majd | 5.09 | 3.90 | -0.77 | 0.87 | 4.93 | 8.69 | 13.56 |
Table 4: Sequence-averaged morphological parameters for BNP3029-DNA using the uncorrelated snap shots from five MD simulations (total length of MD run is 140 ns, RESP charges were used for ((C6H5)(CH2)3C≡N)2Ptgg). 2K0V is unplatinated DNA from NMR studies. The 2K0V numbers highlighted in green are within Mean ± SD values from the MD run. SD is the standard deviation. Min and Max are the minimum and maximum values obtained for that parameter from the MD run. 25%, Median (50%) and 75% are the respective percentiles between Min and Max. tbend is total bend. Ax-bend is a re-rendering of the total bend using the local base-step data. Helical-Rise and Helical-Twist correspond to the translation and rotation of successive base pairs along and around the helical axis. Backbone angles are presented separately for “W” (platinated strand) and “C” (complementary strand). Under Morphological Parameter column, min and maj refer to minor and major grooves and w and d stand for width and depth.
Table 2 compares the sequence-averaged morphological data for the cisplatin-DNA adduct from MD runs with the sequence-averaged data from the crystal structure (3LPV, both molecules A and B) and NMR data (1A84), and Table 3 compares the sequence-averaged morphological data for oxaliplatin-DNA adduct from MD runs and crystal (1IHH) and NMR (2K0U) studies. NMR Data on B-DNA (2K0V) is included for comparison purposes in Tables 2 and 3. As seen in Tables 2 and 3, most of the parameters from x-ray and NMR studies for cisplatin-DNA and oxaliplatin-DNA adducts are within one SD of the mean from the MD runs with total bend angle for cisplatin-DNA adduct from NMR data (83.2°;) being one of the exceptions. Since there are no crystal or NMR structure determinations of the BNP3029-DNA adduct, Table 4 compares the sequence-averaged morphological data for BNP3029-DNA adduct from MD runs with the corresponding data for B-DNA from NMR studies (2K0V). 3S shows the average total bend (tbend) angle of DNA from B-DNA, cisplatin-, oxaliplatin-, and BNP3029-DNA adducts and indicates that the tbend angle for BNP3029-DNA adduct is slightly lower compared to the tbend value in cisplatin- and oxaliplatin-DNA adducts. Also, the tbend for cisplatin-DNA is much smaller than the NMR determined value of 83.2°; and is closer to the x-ray value.
Can we track the global changes in platinated-DNA to individual base-pairs?
The platination changes the morphology of DNA, and the sequence-averaged MD data from Tables 1-4 summarizes a range of values for various parameters. In order to pinpoint if a particular parameter from platinated-DNA is statistically significantly different from the same parameter in B-DNA, we concentrated on eight bases (T3-T10, excluding the two end bases from 5’- and 3’- side of platination) and compared the 42 parameters for each of the 8 bases using the KS statistics. The KS statistics for the 8 bases are shown as heat-maps for B-DNA and for each platinated-DNA (Figures 5-8). As mentioned in Section 2.5, a given morphological parameter, for a given base in platinated-DNA, is assigned as significantly different compared to the same parameter for the same base in B-DNA if at least 4 or more cells are brown in color.
Figure 5:The heat map for B-DNA from MD runs. The heat-map was presented with four colors: background light pink color refers to Kolmogorov-Smirnov (KS) deviation for a given parameter from that of normal B-DNA in the range of 0≤p<0.8 (p is the probability deviation, as p gets closer to 1, then higher the probability of deviation); dark yellow color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.8≤p<0.9; orange color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.9≤p<0.95; and the brown color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.95≤p<1.0. For each of the 42 morphological parameters, each base (T3 to T10) contains 5 cells representing the five MD runs (either 10 ns or 60 ns). A given morphological parameter, for a given base in platinated-DNA, was assigned as significantly different compared to the same parameter for the same base in B-DNA if at least 4 or more cells were brown in color. For each cell the average data were generated using the uncorrelated samples from MD runs.
Figure 6: The heat map for cisplatin-DNA from MD runs. The heat-map was presented with four colors: background light pink color refers to Kolmogorov-Smirnov (KS) deviation for a given parameter from that of normal B-DNA in the range of 0≤p<0.8 (p is the probability deviation, as p gets closer to 1, then higher the probability of deviation); dark yellow color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.8≤p<0.9; orange color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.9≤p<0.95; and the brown color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.95≤p<1.0. For each of the 42 morphological parameters, each base (T3 to T10) contains 5 cells representing the five MD runs (either 10 ns or 60 ns). A given morphological parameter, for a given base in platinated-DNA, was assigned as significantly different compared to the same parameter for the same base in B-DNA if at least 4 or more cells were brown in color. For each cell the average data were generated using the uncorrelated samples from MD runs using the RESP charges for cisplatin-DNA.
Figure 7: The heat map for oxaliplatin-DNA from MD runs. The heat-map was presented with four colors: background light pink color refers to Kolmogorov-Smirnov (KS) deviation for a given parameter from that of normal B-DNA in the range of 0≤p<0.8 (p is the probability deviation, as p gets closer to 1, then higher the probability of deviation); dark yellow color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.8≤p<0.9; orange color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.9≤p<0.95; and the brown color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.95≤p<1.0. For each of the 42 morphological parameters, each base (T3 to T10) contains 5 cells representing the five MD runs (either 10 ns or 60 ns). A given morphological parameter, for a given base in platinated-DNA, was assigned as significantly different compared to the same parameter for the same base in B-DNA if at least 4 or more cells were brown in color. For each cell the average data were generated using the uncorrelated samples from MD runs using the RESP charges for oxaliplatin-DNA.
Figure 8: The heat map for BNP3029-DNA from MD runs. The heat-map was presented with four colors: background light pink color refers to Kolmogorov- Smirnov (KS) deviation for a given parameter from that of normal B-DNA in the range of 0≤p<0.8 (p is the probability deviation, as p gets closer to 1, then higher the probability of deviation); dark yellow color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.8≤p<0.9; orange color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.9≤p<0.95; and the brown color refers to KS deviation for a given parameter from that of B-DNA MD run in the range of 0.95≤p<1.0. For each of the 42 morphological parameters, each base (T3 to T10) contains 5 cells representing the five MD runs (either 10 ns or 60 ns). A given morphological parameter, for a given base in platinated-DNA, was assigned as significantly different compared to the same parameter for the same base in B-DNA if at least 4 or more cells were brown in color. For each cell the average data were generated using the uncorrelated samples from MD runs using the RESP charges for BNP3029-DNA.
T3 base: T3 base in all three platinated-DNA adducts showed significant deviations in ax-bend angle (which is a re-rendering of total bend using the local base-step data) and cisplatin- and BNP3029- DNA adducts also differed significantly in base-pair (T3-A22) axis parameter- tip (θ) compared to the value for the T3 base in B-DNA.
C4 base: Many morphological parameters of C4 base showed significant deviation from that of B-DNA as it approaches platinated-gg such as: intra base-pair (C4-G21) parameter- buckle (κ); base-pair (C4- G21) axis parameter- inclination (η); ax-bend angle; and major groove width and depth for all three platinated-DNA adducts. Base-pair (C4- G21) axis parameter- tip (θ) and base-pair step (C4/T5) parameter- slide (Dy) were different with cisplatin-DNA and oxaliplatin-DNA adducts compared to that in B-DNA. Also, the cisplatin-DNA adduct showed significant deviations in base-pair step (C4/T5) parameters- roll (ρ) and twist (Ω) angles and backbone angle- delta (δ) on the C-strand (no platinum adduct) from the corresponding values in B-DNA. Whereas the BNP3029-DNA adduct significantly differed in the backbone chi (χ) angle on the C-strand from the corresponding value in B-DNA.
T5 base: Since T5 base is next to the 5’-platinated g6, several morphological parameters of T5 base showed significant deviation from those of the T5 in B-DNA. These include: base-pair (T5-A20) axis parameters- inclination (η), and tip (θ); ax-bend angle; base-pair step (T5/g6) parameters- tilt (τ) and twist (Ω) angles; and major and minor groove widths were different in all the three platinated-DNA adducts compared to those in B-DNA. Other significantly different parameters include: intra base-pair (T5-A20) parameter- stagger (Sz) and base-pair (T5-A20) axis parameter- Ydisp (dy); and backbone angles- zeta (ζ) and the sugar phase (P) on the W-strand for cisplatin- DNA and oxaliplatin-DNA adducts; backbone angle alpha (α) on the C-strand; and major and minor groove depths in cisplatin-DNA adduct; intra base-pair (T5-A20) parameter- propel (ω); base-pair step (T5/g6) parameter- roll (ρ); and backbone angle delta (δ) on the W-strand in oxaliplatin-DNA adduct; and intra base-pair (T5-A20) parameter- propel (ω); base-pair step (T5/g6) parameters- shift (Dx) and slide (Dy); and backbone angles delta (δ) and sugar pucker (P) on the W-strand and major and minor groove depths in BNP3029-DNA compared to the values in B-DNA.
g6 base: As the base that is platinated on the 5’-side, several of the morphological parameters of g6 base showed significant deviation from those of G6 in B-DNA in all the three platinated-DNA. These include: intra base-pair (g6-C19) parameter- propel (ω); base-pair (g6-C19) axis parameters- inclination (η), and tip (θ); ax-bend angle; base-pair step (g6/g7) parameters- shift (Dx), rise (Dz), roll (ρ), and twist (Ω); the backbone angles- beta (β) on the C-strand; and both major and minor groove widths and depths. An increased roll angle between g6/g7 base-step from a typical value of 0°; (in B-DNA) leads to changes in groove dimensions. For example, a positive roll angle opens up the minor groove (increases minor groove width) and consequently narrows the major groove. Because of the increased roll angle in all of the three platinated-DNAs, the platinated-DNAs do not have significant deviations in the major and minor groove widths and depths. Other significantly different parameters include: intra base-pair (g6-C19) parameters- shear (Sx) and opening (σ); base-pair step (g6/ g7) parameter- slide (Dy); backbone angles delta (δ), zeta (ζ), chi (χ) and sugar phase (P) on the W-strand for cisplatin-DNA adduct; intra base-pair (g6-C19) parameter- shear (Sx) and backbone angles delta (δ), zeta (ζ), and sugar phase (P) on the W-strand and sugar pucker amplitude (τmax, the sugar puckering amplitude measures the extent of deviations of the five torsion angles in the five-membered ring from zero) on the C- strand for oxaliplatin-DNA; and intra base-pair (g6- C19) parameter- buckle (κ); base-pair step (g6/g7) parameter- slide (Dy); and backbone angles epsilon (ε) and chi (χ) on the W-strand for BNP3029-DNA adduct.
g7 base: As the base that was platinated on the 3’-side, again, several morphological parameters of the g7 base showed significant deviation from those of B-DNA in all three platinated-DNAs. These include: intra base-pair (g7-C18) parameter- buckle (κ); base-pair (g7- C18) axis paramters- Xdisp (dx), Ydisp (dy), inclination (η), and tip (θ); ax-bend angle; base-pair step (g7/T8) parameters- slide (Dy) and rise (Dz); the backbone angles: beta (β), and sugar phase (P) on the W-strand; and minor groove widths and depths and major groove depth. Other significantly different parameters include: intra base-pair (g7-C18) parameters- shear (Sx), propel (ω), and opening (σ); basepair step (g7/T8) parameter- roll (ρ) and backbone angles delta (δ) on the W-strand and alpha (α), delta (δ), zeta (ζ), sugar phase (P) and amplitude on the C-strand for cisplatin-DNA adduct; intra base-pair (g7-C18) parameter- propel (ω); base-pair step (g7/T8) parameter- tilt (τ); sugar pucker amplitude on the W-strand and backbone angles alpha (α), delta (δ), zeta (ζ) and sugar phase (P) and sugar pucker amplitude on the C-strand for oxaliplatin-DNA adduct; intra base-pair (g7-C18) parameter- opening (σ); base-pair step (g7/T8) parametersshift (Dx) and twist (Ω); and backbone angle delta (δ) and amplitude on the W-strand for the BNP3029-DNA adduct.
As mentioned above and as seen in Figure 8, there are fewer dark brown cells for C4, T5, g6 and g7 base morphological parameters for the BNP3029-DNA adduct compared to more dark brown cells for cisplatin- and oxaliplatin-DNA adducts for the same bases (Figures 6 and 7).
T8 base: Since the 3’-side hydrogens on ammine in cisplatin or the hydrogens on DACH NH2 in oxaliplatin interact in a hydrogenbond fashion with the O4 atom of T8 and O6 atom of g7, both of these adducts (cisplatin-DNA and oxaliplatin-DNA) showed significant changes in the T8 base backbone angles such as zeta (ζ), chi (χ) and sugar phase (P) on W-strand and delta (δ), epsilon (ζ), chi (χ) and sugar phase (P) on C-strand compared to those in B-DNA. Since oxaliplatin- DNA had more hydrogen-bond interactions than cisplatin-DNA, the T8 base in oxaliplatin-DNA also showed changes in base-pair step (T8/ C9) parameters- shift (Dx) and roll (ρ); and sugar pucker amplitude on W-strand and zeta (ζ) on C-strand compared to B-DNA. Both major and minor groove width and depths in cisplatin-DNA and minor groove width and depths in oxaliplatin-DNA adducts significantly differed compared to those in B-DNA. All three platinated-DNA adducts have deviations in intra base-pair (T8-A17) parameter- propel (ω); base-pair (T8-A17) axis parameters- Ydisp (dy), and inclination (η); and ax-bend from B-DNA. Cisplatin-DNA and oxaliplatin-DNA have significant deviations in base-pair (T8-A17) axis parameter- tip (θ); and cisplatin-DNA adduct alone has deviations in intra basepair (T8-A17) parameter- buckle (κ); and backbone angle delta (δ) on W-strand and alpha (α) on C-strand; whereas BNP3029-DNA has significant deviations in zeta (ζ) on W- strand and in major and minor groove widths compared to B-DNA. Overall, compared to cisplatin- DNA and oxaliplatin-DNA, BNP3029-DNA had fewer deviations from B-DNA at the T8 base.
C9 base: As the bases move away from platinum adduct on the 3’- side, fewer number of morphological parameters in platinated-DNA deviate from those in B-DNA, as seen with the C9 base. For cisplatin- DNA, oxaliplatin-DNA and BNP3029-DNA adducts, base-pair (C9- G16) axis parameter- tip (θ); ax-bend angle; and major groove width and depth showed significant deviation from those of B-DNA. Other deviations include: base-pair (C9-G16) axis parameters- Xdisp (dx), Ydisp (dy) and inclination (η) for oxaliplatin-DNA adduct; and basepair (C9-G16) axis parameters- Ydisp (dy) and inclination (η) and backbone angle alpha (α) on the C-strand for cisplatin-DNA.
T10 base: Since T10 is even farther from the platinum adduct, it displayed fewer deviations in its morphological parameters in platinated-DNA compared to B-DNA. Ax-bend from all three platinated adducts showed significant deviations from that of B-DNA. Other deviations include: base-pair (T10-A15) axis parameter- tip (θ) and major groove width and depth for oxaliplatin-DNA adduct; and base-pair (T10-A15) axis parameter- tip (θ) for cisplatin-DNA adduct.
Overall for most of the parameters, the KS plots indicate that BNP3029-DNA adduct has less of a deviation from those of the B-DNA than either ciplatin-DNA or oxaliplatin-DNA adducts. Next, the tetranucleotide- T5g6g7T8 is used to explain the conformational preference of DNA (B-DNA vs A-DNA) in the vicinity of platination for the three platinated-DNA adducts by monitoring the key parameters such as roll, slide, twist and rise; delta, zeta, and chi angles; inclination; major and minor groove dimensions; sugar pucker preferences; and BI and BII DNA conformers in the vicinity of platination.
Base-Step parameter changes: roll, slide, twist and rise
The base-step parameter roll angle (ρ) is defined as the deformation along the base-pair long axis and describes the groove bending. For example, a positive roll angle (as seen here with platinated-DNA adducts) opens up the minor groove (increases the width of minor groove) and consequently narrows the major groove. Crystallographic and solution studies indicate that platination of DNA by cisplatin and oxaliplatin induces a significant (positive) roll angle change at the site of platination between g6/g7 base-step compared to that in B-DNA. Figure 9 shows the changes in roll angle for T5/g6, g6/g7, g7/T8 and T8/C9 base-steps obtained from the MD studies. As seen from crystal and NMR studies, the average roll angle from MD studies between g6/ g7 base-step is significantly different in platinated-DNA compared to the G6/G7 base-step roll angle in B-DNA. Of all the three platinated- DNA adducts, BNP3029-DNA showed less of a roll angle between g6/ g7 base-step compared to the other two platinated-DNA adducts.
The base-step parameter slide (Dy) is defined as the translation along the base-pair long axis (relative displacement of successive base pairs) which is around -1.4 Å for A-DNA and around 0 for B-DNA. Figure 4S shows the average value from the MD data in slide values for the above four base-steps. Of all the 4 base-steps, the g6/g7 basestep has the largest negative slide value. Since A-form DNA has negative slide values, the large negative slide values for g6/g7 base-step indicate a change to A-form. Another base-step parameter that could potentially indicate a change from A- to B-form is the twist angle (Ω, helical rotation between successive base-pairs) which is around 30°; for A-DNA and around 36°; for B-DNA. Figure 5S shows the average twist values for the tetranucleotide T5/g6, g6/g7, g7/T8 and T8/C9 basesteps. Again, from Figure 5S, the g6/g7 base-step in platinated-DNA has lower twist values indicating a form change in DNA. Of the three platinated-DNA, BNP3029-DNA has twist values closer to B-DNA compared to the other two platinated-DNA. Figure 6S shows the average rise (Dz, translational motion of successive base-pairs along the z-axis) values, which is around 2.6 Å for A-DNA and around 3.4 Å for B-DNA, for the tetranucleotide T5/g6, g6/g7, g7/T8 and T8/C9 base-steps. From Figure 6S, the g6/g7 base-step in platinated-DNA has higher rise values compared to the rise value for G6/G7 base-step in B-DNA.
Changes in backbone angles: delta, zeta, and chi
Backbone angles that can distinguish A- and B-DNA morphologies are delta (δ), zeta (ζ) and chi (χ). Delta value for canonical A-DNA is around 79°; and for B-DNA is around 143°;. Figure 7S shows both the W- and C- strand average delta (δ) values for the tetranucleotide T5g6g7T8. As seen earlier with rise and twist, the delta values for T5 and g6 base are low in cisplatin- and oxaliplatin-DNA compared to values observed in B-DNA whereas BNP3029-DNA values are closer to that of B-DNA.
The zeta (ζ) value for canonical A-DNA is around -75°; and B-DNA is around -161°;. Figure 8S shows both the W- and C- strand average zeta (ζ) values for the tetranucleotide T5g6g7T8 in platinatedand B-DNA. T5, g6 and g7 bases show more A-like character in platinated-DNA. The chi (χ) value for canonical A-DNA is around -157°; and B-DNA is around -98°;. Figure 9S shows (for both W- and C- strand) the average chi (χ) values for the tetranucleotide T5g6g7T8 in platinated- and B-DNA. Notable changes in chi values are seen for A17 base of cisplatin- and oxaliplatin-DNA. This deviation may be a result of increased hydrogen bonding between cisplatin and oxaliplatin carrier ligand on the 3’- side of platinum adduct with the T8 base. Also, the chi value for g6 base in BNP3029- is closer to B-DNA value compared to the value seen in cisplatin- and oxaliplatin-DNA for the same base. Overall, BNP3029-DNA has fewer number of bases deviant from B-DNA in delta (δ) and chi (χ) angle compared to cisplatin-DNA or oxaliplatin-DNA.
Changes in inclination
Base-pair rotation about the short-axis is defined as inclination (η). This angle is 0°; for B-DNA whereas it is around 20°; for A-DNA. Figure 10S shows the average inclination (η) values for the tetranucleotide T5g6g7T8 in platinated- and B-DNA. Based on Figure 10S, the inclination values for the 4 bases in platinated-DNA are significantly different from B-DNA values.
Changes in major and minor groove dimensions
Figure 10 shows the average widths for minor (panel A) and major (panel B) grooves for the tetranucleotide T5g6g7T8 in platinated- and B-DNA. From Figure 10 (panel A) it is clear that the minor groove has widened considerably for g6 and g7 bases in all three platinated-DNA compared to the minor groove width for these bases in B-DNA. In addition, T5 and T8 bases in all three platinated-DNA also show widening of the minor groove compared to the same bases in B-DNA to accommodate the platinum adduct. As seen in Figure 10 (panel A), for all these 4 bases, the widening of the minor groove for BNP3029- DNA is lower compared to cisplatin-DNA and oxaliplatin-DNA indicating a more B-like morphology. Increased minor groove width will be followed by a decrease in major groove width (Figure 10, panel B). Again, BNP3029-DNA shows slightly increased major groove widths in line with a more B-like DNA compared to the other two platinated-DNA. As seen from the experimental studies, the minor groove depth becomes shallower and the major groove depth becomes deeper for platinated-DNA. Based on Figure 11S (panels A and B), BNP3029-DNA shows more B-DNA like trends in minor and major groove depths compared to the other two platinated-DNA.
Sugar Pucker
Figure 11 shows the percent (%) A-like sugars (average value) for W-strand (panel A) and C-strand (panel B) for the tetranucleotide T5g6g7T8 in platinated- and B-DNA. As seen from Figure 11 (panel A, the W-strand which has the platinum attached to the two ggs), the sugar on base T5 is less A-like in the case of BNP3029-DNA compared to cisplatin- and oxaliplatin-DNA. Similarly, the sugar on base g6 is more A-like in both cisplatin- and oxaliplatin-DNA whereas in BNP3029- DNA the A-like character on g6 base is reduced pronouncedly. On the complementary strand (C-strand, Figure 11, panel B) bases C18 (opposite to g7) and A17 (opposite to T8) have substantially higher A-like sugars in cisplatin- and oxaliplatin-DNA compared to BNP3029- DNA. This may be due to the two hydrogen-bonds found in cisplatinand oxaliplatin-DNA (The first one is between the O6 atom of g7 base and the NH2 hydrogens of DACH (or all three NH3 hydrogens in the case of cisplatin) and the second one is between the O4 atom of T8 base and the NH2 hydrogens of DACH (or all three NH3 hydrogens in the case of cisplatin)). Overall, BNP3029 platinated-DNA has less of an A-like character in its base sugars compared to the base sugars in cisplatin-DNA or oxaliplatin-DNA.
Figure 11: Averaged percent (%) occupation of A-like sugars (combining C3’-endo, C4’-exo and C2’-exo sugar puckers, panel A: W-strand that has the platinum adduct and panel B: C-strand, the complementary starnd) for the 12-mer DNA (5’-CCTCTggTCTCC-3’) from the platinated-DNA and B-DNA. The average data were generated using the uncorrelated samples from five MD runs using the RESP charges for the platinated-DNA. Panels A and B contain data for Bases 3 to 10 only.
BI and BII DNA Conformers
Experimentally B-DNA is known to exist in 80% BI conformer and 20% in BII conformer [24,25]. These two conformers arise due to the population differences in epsilon (ε) and zeta (ζ) backbone angles (ε-ζ<0 leads to BI and ε-ζ>0 leads to BII conformer). As seen from Figure 12S (panel A, the W-strand which has the platinum bonded to the two ggs), most of the time, BI conformation is dominant for B-DNA or cisplatin- DNA, oxaliplatin-DNA or BNP3029-DNA except (i) in B-DNA the BI conformation for g6 base is only 80%, with the rest being BII and (ii) the T8 base showed less of BI conformation in cisplatin-DNA, oxaliplatin- DNA and BNP3029-DNA. Again, a plausible explanation for the lower occupancy of BI conformer in oxaliplatin-DNA or cisplatin-DNA is due to the enhanced hydrogen-bonding between the O4 atom of T8 with one of the NH2 hydrogens on DACH in oxaliplatin or with the NH3 hydrogens of cisplatin. On the complementary strand (C-strand, Figure 12S, panel B) base A20 (opposite to T5) in B-DNA has lower BI conformation and the same base in cisplatin-DNA, oxalipaltin-DNA and BNP3029-DNA has even lower BI conformation compared to the B-DNA.
Changes in backbone angles: alpha and gamma
Since the current simulations employed parm99 forcefield that is modified with parmbsc0 forcefield, all the bases in B-DNA, cisplatin-DNA, oxaliplatin-DNA and BNP3029-DNA maintained a g- (-60)/ g+ (+60) conformation for α / γ angles respectively in both strands with the exception of (i) the base G6 (W-strand) in B-DNA; (ii) the base T8 (W-strand) in cisplatin; (iii) the base C9 (W-strand) in oxaliplatin; and (iv) the base A20 (C-strand, opposite to T5) and the base G21 (C-strand, opposite to C4) in BNP3029-DNA.
Since the discovery of cisplatin’s antitumor properties, many platinum analogues have been made and tested in clinical trials yet few have received FDA appoval. Several studies have focused on obtaining the activation energy barriers for the attack of nucleophiles on cisplatin or the monoaquated cisplatin to understand the platinum analogue’s reactivity [6]. Also, crystal structure and NMR studies have been directed at platinum-DNA adducts to understand the morphological changes in DNA by platinum adduct formation [14-18]. These studies indicated that guanine is the primary site of attack for monoaquated cisplatin over adenine. Platination of two adjacent guanines (gg) bends DNA and induces changes in the roll angle between the two guanines (gg) along with other morphological changes. It has also been reported that certain platinum-DNA adducts did not deform DNA as seen with cisplatin-DNA and oxaliplatin-DNA adducts.
The elaborate molecular dynamics simulations conducted in this study on B-DNA and platinated-DNA (cisplatin, oxaliplatin or BNP3029) revealed several interesting details: (i) overall RMSDs for DNA are in the expected range [28]; (ii) substituted cyano groups (carrier ligand) in BNP3029 occupy the entire major groove area near g6g7 bases (which are platinated) compared to the limited space occupied by cisplatin (NH3s) or oxaliplatin (DACH, Figure 3) carrier ligands. This may have implications for the downstream cellular events similar to the recent studies that indicated the presence of bulky arylamine substitutions in the major groove prevent translesion synthesis [29,30]; (iii) Watson-Crick base-pair hydrogenbonds are more or less maintained in all three platinated-DNAs with the exception of the hydrogen-bond between the O2 atom of g6 and N2(H21) atom of C19 which was characterized by a drop in occupancy in all three platinated-DNAs. The hydrogen-bond between O4 atom of T8 and N6(H61) atom of A17 also showed a drop in occupancy with cisplatin-DNA and oxaliplatin-DNA adducts. The drop in occupancy for this hydrogen-bond in cisplatin-DNA and oxaliplatin-DNA is due to the enhanced hydrogen-bonding between the O4 atom of T8 with the NH2/NH3 hydrogens of oxaliplatin and cisplatin respectively. This type of hydrogen-bond is not possible with BNP3029-DNA; (iv) MD simulations on platinated-DNA indicated that the hydrogen-bonds are seen between the ligand hydrogens (cisplatin and oxaliplatin only) with the 3’-side bases of DNA (O6 atom of g7 and O4 atom of T8) only but not with any 5’-side bases.
The sequence-averaged (across 8 base-pairs) morphological data from the elaborate MD studies on platinated-DNA (platinated with cisplatin, oxaliplatin or BNP3029) is very similar to the sequenceaveraged data from the MD studies on B-DNA indicating globally these DNA structures are similar. Local deformations are observed with g6g7 (platinated guanines) and the nearby bases (T5 and T8). One of the distinct deformations of DNA by platination is the increased roll angle between g6g7. A consequence of the increased roll angle between g6g7 is the widening of the minor groove and a consequent narrowing of the major groove. The roll angle between g6g7 is smaller for BNP3029-DNA, compared to the other two platinated-DNAs with cisplatin and oxaliplatin, and as a result the groove widths and depths in BNP3029-DNA are qualitatively closer to B-DNA than the other two platinated-DNA. A similar trend is also seen in the BNP3029-DNA sugar puckers (especially the tetranucleotide T5g6g7T8,) with T5, g6, C18, and A17 showing more B-like sugars compared to platinated- DNA with cisplatin and oxaliplatin. The total bend angle of DNA is also slightly lower in the case of BNP3029-DNA as compared to the other two platinated-DNAs. Overall, BNP3029-DNA has more B-DNA like character compared to cisplatin-DNA or oxaliplatin-DNA, and this could indicate that substituted cyano platinum agents may have distinct reactivity properties. Cytotoxicity studies in a variety of wildtype and resistant human cancer cell lines indicate that BNP3029 is more potent in comparison to cisplatin and oxaliplatin [6].
Herein, morphological changes in DNA upon platination by cisplatin, oxaliplatin and BNP3029, studied using molecular dynamic simulation (MD) studies, are reported. A combined total of ~250 ns of MD simulations were carried out for each of the three platinum analogues using a 12-mer DNA oligonucleotide sequence (5’-CCTCTggTCTCC-3’; gg is the site of platinum adduct formation) employing electrostatic potential or restrained electrostatic potential charges on platinum-gg adduct. Morphological data from platinated- DNA were compared with those from B-DNA (100 ns MD data). Some of the changes observed in all the three platinated-DNAs include: 1) increased total bend angle; 2) increased roll angle between adjacent guanines (gg); and 3) changes in the groove dimensions compared to B-DNA. Ammine hydrogens in cisplatin and oxaliplatin participated in hydrogen-bond interactions with the DNA bases g7 and T8 (bases on the 3’-side of the platinum adduct). The substituted cyano ligands of BNP3029 occupy the entire major groove area near g6 and g7 which may have implications for downstream cellular events. The two phenyl rings of BNP3029 adopt either a T-shaped or a Y-shaped arrangement (~40% of the time during the MD runs). Based on Kolmogorov-Smirnov statistical analyses on the DNA morphological data, platinated-DNA with BNP3029 showed less of a deviation from B-form as compared to the changes seen with platinated-DNA with cisplatin or oxaliplatin. These unique characteristics of BNP3029 and BNP3029-DNA may contribute to the previously reported more potent activity by BNP3029 in a variety of human cancer cell lines including cell lines traditionally resistant to cisplatin and oxaliplatin [6].