Journal of Cancer Research and Immuno-Oncology
Open Access
ISSN: 2684-1266
ISSN: 2684-1266
Research Article - (2023)Volume 9, Issue 3
Background: Myeloid or lymphoid progenitors in the bone marrow can develop into leukemia, which is clonal cancer. Acute Myeloid Leukemia (AML) developed as a result of somatic mutations in the precursor cells of the myeloid lineage along with transcriptome dysregulation of bone marrow infiltration, which resulted in the generation of immature myeloid cells (blasts) and disruption of normal hematopoiesis. Current research has demonstrated the metastasis potential of a certain subset of microRNAs. As a result, miRNA downregulation at the transcriptional level can reduce the possibility of metastasis. The purpose of this work is to analyze miRNA precursor targeting utilizing the CRISPR-C2c2 (Cas13a) method.
Results: crRNAs designed for miR-301b and miR-21 has a very high structural similarity with binding energy to the state observed in the normal condition.
Conclusion: Sequence based evaluation of crRNAs intended for RNA level editing is insufficient; simulation and molecular docking investigations should also be carried out for improved accuracy.
AML; Computational analysis; CRISPR/C2c2; Metastasis; Lymphoid progenitors; Acute myeloid leukemia
In terms of genesis, clinical presentation, and prognosis, Acute Myeloid Leukemia (AML) is a diverse hematological malignancy [1]. Clinical results in AML patients are typically subpar despite current treatment [2]. Myeloid or lymphoid progenitors in the bone marrow can develop into leukemia, which is a clonal cancer. AML emerged from somatic mutations in myeloid lineage precursors along with transcriptome dysregulation of bone marrow infiltration, which led to the production of immature myeloid cells (blasts) and interruption of regular hematopoiesis. AML is identified by high blast counts in the blood, where a presentation of 20% or more blasts is deemed to be AML [3]. The susceptibility of AML sufferers to life-threatening infectious headaches is enormously stated as its miles the important motive of morbidity and mortality on this group [4]. miRNAs are small non-coding RNAs (18 nucleotides-24 nucleotides) that perform post-transcriptional regulation of mRNAs, mainly playing an inhibiting role when binding to the mRNA 3 untranslated region, ultimately impeding their translation or leading to degradation [5]. miRNAs regulate several biological processes, including cellular differentiation, proliferation, and apoptosis; thus, they can be easily found in blood samples and are easy to obtain and minimally invasive [5]. miRNAs are also essential for cancer progression due to reported observations that they play critical roles in regulating cancer signaling mechanisms, enhancing several factors, including tumor growth, angiogenesis, and metastasis [6,7]. Despite numerous discoveries in the CRISPR/Cas system, its application in diagnostics and gene therapy has only been developed in the last few decades. In 2012, it was reported that CRISPR nucleases can be programmed simply by changing the guide RNA (gRNA) sequence, which is extremely useful for genome editing [8]. In 2013, scientists succeeded in editing a variety of cell types using a specific type II CAS protein called Cas9 [9]. In 2015, two CRISPR/Cas type V proteins, Cas12a and Cas13a, were discovered for genome editing and were originally named Cpf1 and C2c2, respectively [10-12]. Cas13a (known as C2c2), another novel RNA-guided system with the ability to target RNA, has recently been discovered [13]. After recognizing and binding to the target RNA, Cas13a will activate the capacity of collateral cleavage to the untarget RNAs. However, the collateral cleavage activity of this system was not identified in eukaryotic species and its underlying mechanism was poorly understood [14]. So far, this type of CRISPR based RNA targeting tool has already been used in biomedical applications, such as the detection of specific sequences of patient viral RNA or tumor circulating RNA [15]. The RNA-targeting gene editing systems have great potential in the treatment of cancer and malignant diseases by manipulating critical RNA molecules (both mRNAs and non-coding RNAs such as microRNAs, lncRNAs, etc.) [16]. In this study, using bioinformatics software and targeting microRNA precursors involved in Acute Myeloid Leukemia (AML) metastasis, a new method for post-transcriptional regulation of this cancer was proposed. Also, in order to increase the accuracy and specificity of the technique proposed in this study (CRISPR-C2c2), first analyzes were performed at the design level of the target crRNA and then structural studies were performed using structural bioinformatics methods. The help of bioinformatics tools, especially the CRISPR-C2c2 system, can have a significant J Cancer Res Immunooncol, Vol.9 Iss.3 No:1000181 2 impact on reducing the cost, time and frequency of trial and error of researchers.
The precursor miRNA sequences of the target were first downloaded from the mirBase server in Fasta format. Targeting adult miRNAs was not acceptable due to their short size and the possibility of creating off-targets. Therefore, miRNAs involved in metastasis were controlled in the precursor stage (Table 1).
| miRNA | Precursor |
|---|---|
| hsa-miR-301b | GCCGCAGGUGCUCUGACGAGGUUGCACUACUGUGCUCUGAGAAGCAGUGCAAUGAUAUUGUCAAAGCAUCUGGGACCA |
| hsa-miR-21 | UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA |
Table 1: miRNAs involved in precursor.
The CRISPR-RT (RNA Targeting) server was used to design and model the target crRNAs. CRISPR-RT is an algorithm that provides the design of crRNA for the CRISPR-C2c2 technique. This system provides commanding a wide range of parameters and crRNA design to be performed with the most accuracy. These parameters can be the length of the complementary region of crRNA, PFS, and the number of erroneous pairing or gaps acceptable by this system. After specifying and setting the desired parameters, the CRISPR-RT system presents a list of crRNA sequences with different characteristics based on the input data for the expected target sequence. By setting the required parameters in the CRISPR-RT server, candidates based on the input RNA sequence and accurate search algorithms of non-target sites for each input sequence in transcriptome and genome were presented. Based on the results of the CRISPRT-RT server, the appropriate crRNAs were selected by considering the parameters of the start and end site of the desired crRNA, GC%, the lowest number of non-target sites in the genome and transcriptome, which indicates the designed sequence specificity.
In fact, the designed crRNAs are used in a structure known as scaffold. In other words, the part designed by CRISPR-RT is used to target the target area and the scaffold part is used to activate the C2c2 enzyme due to its second and third structure. The scaffold used in this study is the proposed structure of the algorithm used in CRISPR-RT, and this sequence with the ring stem structure has been confirmed and presented based on previous research. In accordance with this structure and the designed part and the target recipient, the three dimensional structure of the crRNA was performed.
The crystallographic structure of C2c2 enzyme (cas13a) was downloading from a protein database with 5wtj code. HDOCK server was used to perform molecular docking between RNAs simulated with C2c2 enzyme (cas13a). This server was used for molecular docking of protein-protein, protein-DNA, and protein-RNA. HDOCK uses global docking to create complexes. Therefore, you do not need to have basic information about the connection range. Docking complexes were analyzed and compared using PyMol.
Simulation crRNAs designed with web tools in order to study the interaction and complexity between crRNAs and C2c2 (Cas13a) enzyme, the sequences of crRNAs designed for the precursors of miRNAs were simulated in 3D. The RNA composer system offers a new user friendly approach to the fully automated prediction of large RNA 3D structures. The method is based on the machine translation principle and operates on the RNA FRABASE database acting as the dictionary relating RNA secondary structure and tertiary structure elements. RNA composer works in two modes: interactive mode allows working on one RNA molecule of interest at a time; its use is limited up to 500 nt residues and results in a single 3D-RNA structure model. Batch mode is designed for large-scale automated modeling of RNA structures up to 500 nt residues, based on user defined RNA secondary structures. As an input a set of up to 10 RNA sequences can be used. This mode is available only for registered users.
The crRNAs designed for the selected miRNAs are listed (Table 1 and Figure 1).
| No | miRNA name | Protospacer+PFS (crRNA) | Start | End | GC | Transcript target | Gene target |
|---|---|---|---|---|---|---|---|
| 1 | hsa-mir-301b | GCCGCAGGUGCUCUGACGAGGUUGCACUA | 1 | 29 | 0.64 | 1 | 1 |
| 2 | hsa-mir-21 | GGCAACACCAGUCGAUGGGCUGUCUGACA | 44 | 72 | 0.61 | 2 | 1 |
Table 2: crRNAs designed for miRNA using CRISPR-RT server.
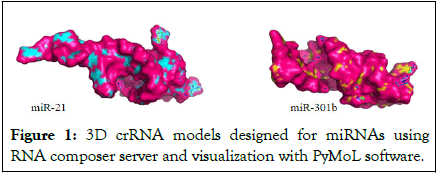
Figure 1: 3D crRNA models designed for miRNAs using RNA composer server and visualization with PyMoL software.
Using the HDOCK server, for each of the target microRNAs, 100 C2c2 (Cas13a) with crRNA simulated docking models were presented. Out of the 100 predicted models, the top 10 models were selected based on energy level (Table 3).
| No | miRNA | Rank | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 | Model 7 | Model 8 | Model 9 | Model 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-mir-301b | Docking score | -326.73 | -319.43 | -317.99 | -290.44 | -290.3 | -282.07 | -281.56 | -279.28 | -275.24 | -272.03 |
| Ligand rmsd (Å) | 207.34 | 117.09 | 179.77 | 120.72 | 117.31 | 122.23 | 144.49 | 122.39 | 118.93 | 115.89 | ||
| 2 | hsa-mir-21 | Docking score | -333.57 | -317.81 | -314.56 | -312.78 | -307.47 | -306.95 | -282.83 | -273.87 | -270.35 | -269.35 |
| Ligand rmsd (Å) | 112.44 | 106.39 | 106.35 | 107.26 | 172.29 | 114.29 | 158.56 | 178.11 | 188.67 | 119.36 |
Table 3: HDOCK server results based on interaction between protein Cas13a (C2c2) and crRNA designed by energy level.
The docking complex of C2c2 enzyme and crRNAs designed were examined and analyzed using Pymol software. Investigation of the formed complexes showed that the crRNA designed for miR-301b has a very high structural similarity with binding energy (model 2: 319.43, model 3: 317.99, model 4: 290.44, model 7: 281.56, model 8: 279.28, model 10: 272.03) to the state observed in the normal condition (Figures 1 and 2). Also, the complexes of crRNAs designed for miR-21 had high structural similarity with binding energy (model 1: 333.57, model 2: 317.81, model 3: 314.56, model 8: 273.78, model 10: 269.35) to the state observed in the normal condition (Figure 3).
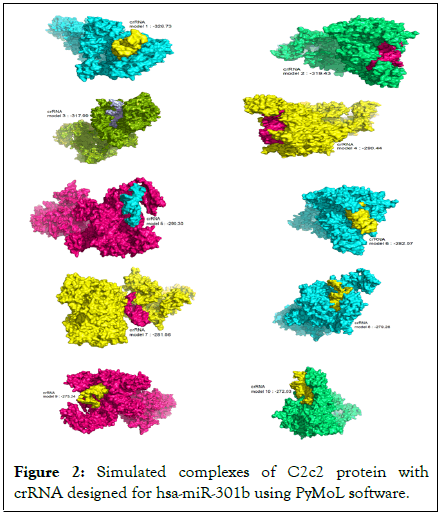
Figure 2: Simulated complexes of C2c2 protein with crRNA designed for hsa-miR-301b using PyMoL software.
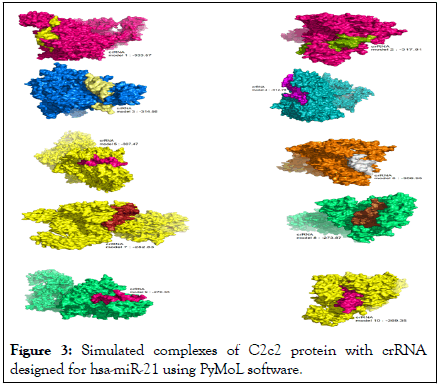
Figure 3: Simulated complexes of C2c2 protein with crRNA designed for hsa-miR-21 using PyMoL software.
A wide range of factors are involved in the occurrence and prognosis of AML. Due to the importance of micro-RNAs in initiation and development of AML, in recent years, extensive studies have shown the significance of miR-301 and miR-21 expression in the leukemia. MiRNA-301b is an onco-miRNA that is over expressed in numerous cancers, including AML, breast and colorectal. In a recent study, Lu et al. revealed that miR-301b targets FOXF2 to give rise to proliferation and inhibit programmed cell death in AML cells through Wnt/β-catenin axis. In line with this, overexpression of miR-301b in bladder cancer cells can lead to down regulation of USP13, and in turn, can inhibit the expression of PTEN (phosphates and tensin homolog), which is one the most decisive tumor suppressor proteins.
Research findings confirm that miR-21 is a significant oncomiRNA which is upregulated in different types of cancers, like breast and colorectal cancers. This miRNA targets several important genes, including PTEN and PDCD4 (programmed cell death protein 4). A recent study in both in vitro and in vivo models of AMLs that are resulted from oncogenic activation of Home box (HOX) transcription factors (HOX based leukemia’s), demonstrated that therapeutic inhibition of miR-21 and miR-196b could cause inhibition of initiation of the leukemia in the models. In accordance with the findings, there are a number of different studies considered the essential roles for the miRNA in AML models. Due to these studies we decided to work on miR-301b-3p and its outcomes on AML by bioinformatics tools. Bioinformatics is an important technique to manage biological big data. A collection of tools and software are used in modern biotechnology, using mathematics and statistics, to gain insight into the biological data and find answers for medical and biological questions.
Replacement treatment with TS-miR has the ability to simultaneously control the expression of numerous cancer-promoting genes, in contrast to traditional molecular targeted medicines and antibody therapies that only affect one molecule. As a result, this therapeutic approach is a next-generation pharmacological modality that can treat highly unmet medical requirements in advanced cancer types.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Data and Graphical Analysis was performed by Omid Moeini. Omid Moeini, Seyed Armit Hosseini, Mahsa Khatibi, Arshia Aliyarzadeh, Amirali Rahmani and Mohammadmatin Nourikhani wrote the manuscript.
Not applicable.
The datasets generated and analyzed during the current study are available in the miRBase: the microRNA database, CRISPR-RT (RNA targeting) server, protein database, HDOCK server, PyMol, RNAComposer and RNA FRABASE repository.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Moeini O, Khatibi M, Hosseini SA, Rahmani A, Nourikhani M, Aliyarzadeh (2023) Computational Analysis of Crrna to Regulate Hsa-Mir-301b-3p and Hsa-Mir-21 Related to Metastasis and Cell Proliferation in Acute Myeloid Leukemia (AML) Using CRISPR/C2c2. J Cancer Res Immunooncol. 9:181.
Received: 30-Apr-2023, Manuscript No. JCRIO-23-23813; Editor assigned: 02-May-2023, Pre QC No. JCRIO-23-23813 (PQ); Reviewed: 16-May-2023, QC No. JCRIO-23-23813; Revised: 05-Jul-2023, Manuscript No. JCRIO-23-23813 (R); Published: 12-Jul-2023 , DOI: 10.35248/2684-1266.23.9.187
Copyright: © 2023 Moeini O, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestrictesed use, distribution, and reproduction in any medium, provided the original author and source are credited.