Research Article - (2023)Volume 8, Issue 2
Background: Since Food and Drug Administration (FDA) approved immune checkpoint inhibitors for the treatment of colorectal cancer in 2017, there have been significant differences in the response of colorectal cancer patients to immune checkpoint inhibitors. In order to achieve better clinical benefits of colorectal cancer patients in immunotherapy, we urgently need to find a biomarker to evaluate whether colorectal cancer patients respond to immunotherapy. In this research, we aimed to explore a biomarker to predict the efficacy of immunotherapy for colorectal cancer patients.
Materials and methods: We collected the expression profiles and clinical information of more than 1800 colorectal cancer patients in 6 cohorts in Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases and meanwhile, we collected 230 immune related signatures by searching literatures. Single sample gene set enrichment analysis was performed on 230 signatures, and the normalized enrichment score of each patient was calculated. Then, the Immune Related Prognosis Score (IRPS) was constructed based on normalized enrichment scores. Finally, an extensive analysis was performed to explore the relationship between the IRPS and immune score, prognostic significance, microsatellite status, immunogenomic factors, cancer genotype and potential immune escape mechanism.
Results: We found that the IRPS can reflect the immune infiltration status of colorectal cancer patients, and it is related to some important immunophenotypic factors, such as neoantigen load and Tumor Mutational Burden (TMB). Further analysis confirmed that patients with the low IRPS had a better response to immune checkpoint inhibitors.
Conclusion: Based on these results, we can conclude that the IRPS may be an available tool for the prediction of immunotherapy efficacy in colorectal cancer patients.
Colorectal cancer; Immunotherapy; IRPS; TMB; Prediction; Efficacy
The Colorectal Cancer (CRC) is one of the most common cancers in the world, and it is also one of the main causes of cancer related death. In 2020, the incidence of CRC ranks fifth worldwide and the mortality is top five in global cancer related death [1]. In China, CRC incidence is higher and CRC ranks second in all cancers. At the same time, CRC cancer is the fourth cancer-related cause of death [2]. At present, the treatment method of CRC includes surgery, chemotherapy, radiotherapy and immunotherapy. Immune Checkpoint Inhibitors (ICIs) are new therapeutic strategies, which targets immune checkpoint molecules, including Programmed Cell Death Protein-1 (PD-1), Programmed Cell Death Ligand Protein-1 (PD-L1) and Cytotoxic T-Lymphocyte-Associated Protein 4 (CTLA4) [3]. In 2017, pembrolizumab (anti-PD-1 antibody) and nivolumab (anti-PD1 antibody) were attained FDA approval for the treatment of metastatic CRC.
The Tumor Microenvironment (TME) is composed of nonmalignant cells, such as Cancer Associated Fibroblasts (CAFs), endothelial cells and pericytes constituting tumor vascular system, immune cells, stromal cells and Extra-Cellular Matrix (ECM), which forms complex interaction with tumor [4-6]. Among the immune cells, except for the common T cells and B cells, TME also includes other immune cells such as dendritic cells, neutrophil, macrophages, and cytotoxic cells, regulatory T cells (Treg), Myeloid Derived Suppressor Cells (MDSCs) and NK cells5. The immunosuppressive cells include mast cells, Treg cells, Myeloid-Derived-Suppressor Cells (MDSCs) and Tumor Associated Macrophages (TAMs) with tumor promoting phenotype. Anti-tumor immune cells of TME mainly comprise T cells, B cells, NK cells and cytotoxic cells [5,7-9]. As a major component of TME, the infiltration level of immune cells contributes to tumor progression and immunotherapeutic response. And the difference of immune cell infiltration level in TME leads to the difference in chemotherapy response in different tumor patients, especially immunotherapy.
In CRC, ICIs only responded to CRC with mismatch repair Deficient And Microsatellite Instability-High (DMMR–MSI-H) and it has no clinical benefit on Mismatch-Repair-Proficient and Microsatellite Instability-Low CRC (pMMR–MSI-L) or Microsatellite Stability (MSS) CRC [10,11]. Now, several reports have indicated that there is an association between the increase of mutation load and the treatment response of anti-CTLA4 antibody or anti-PD-1 antibody [12-14]. Studies have found that neo-antigen load is related to Tumor Mutation Burden (TMB), so neo-antigen load is also related to response [13,14]. Only a subset of CRC patients benefits from immunotherapy, the precise biomarkers to predict immunotherapy efficacy are needed. Therefore, it is essential to confirm biomarkers and screen the dominant populations of ICIs efficacy.
Here, we are committed to establishing an immune signature of CRC to analyze the relationship between immune activities, tumor microenvironment and cancer genotype of CRC. On the basis of these prognosis-related immune signatures, the Immune Related Prognosis Score (IRPS) for CRC was developed, which was found to be related to the overall survival and immuno-phenotypic factors of patients with CRC. Moreover, we found that the IRPS can predict the patient's response to immune checkpoint blockade.
Datasets collection and processing
Gene expression profiles and clinical datasets of CRC were collected from Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) and PubMed. At the same time, downloading the genomic data of TCGA CRC from the Pan-cancer Atlas (https://gdc.cancer.gov/about-data/publications/pancanatlas).
Downloading the raw Chronic Eosinophilic Leukemia (CEL) files generated by Affymetrix in the GEO. The function Robust Multichip Average (RMA) in “affy” of R package was used to process and standardize the CEL files generated by Affymetrix. For the expression value of same gene symbol, we take the gene with the largest expression value.
Finally, more than 1800 tumor samples were obtained from 6 cohorts. GSE39582, GSE17538 and GSE103479 cohorts were used as discovery datasets; At the same time, TCGA, GSE12945 and GSE106584 cohorts were used as validation datasets. The potential batch effect between multicenter datasets was removed by the combat function in the R package “sva” [15].
In addition, we obtained two immunotherapy related cancer cohorts (GSE78220 and IMvigor210) from the GEO database and literature [16]. Melanoma patients in the GSE78220 cohort received anti-PD1 antibody therapy. Bladder cancer patients in the IMvigor210 cohort were treated with inhibitors of PD-L1.
Acquirement of immune-related data
We collected 230 immune related signatures by reading the literatures. The main signatures include 68 gene sets derived from the work of Wolf et al. [17], 34 gene sets from Meta-map [18] ,32signatures were obtained from the ImmuneSigDB (Collection C7 of MSigDB, Broad Institute) [19], 25 signatures were obtained from Bindea [20], 10 oncogenic pathway signatures [21], NK cell related signature [22], Exhausted CD8+T cells signatures [23], APMs gene sets [24] and 85 Th1/IFN-γ gene signature [25]. More details can be seen in supplementary.
Single sample gene set enrichment analysis and immunerelated estimation
The single sample Gene Set Enrichment Analysis (ssGSEA) was performed by using R package “GSVA”, and The Normalized Enrichment Scores (NES) of 230 immune related signatures in each CRC sample were calculated by ssGSEA.
The immune score, tumor purity, stromal score and ESTIMATE score of each CRC patient are calculated on the basis of the NES, which is accomplished by R package “estimate”.
The immune-related prognostic score for colorectal cancer
R package “GSVA” was used to implement ssGSEA for the signature of each sample and calculated the NES so that evaluating the immune infiltration level of immune related signature. Univariate Cox proportional hazards regression model was used to assess the correlation between the NES and overall survival in each sample of 6 CRC cohorts. Then, the fixed effect model in R package “meta” was used to evaluate the overall Hazards Ratio (HR) and P-value.
Finally, the IRPS combined with discovery datasets and immune cell signatures was defined as:
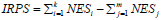
Among them, NESi is the standardized ssGSEA score of the ith immune signature with total HR less than 1, and NESj is the standardized ssGSEA score of the jth immune signature with total HR more than 1.
Statistical analysis
The heatmaps and other plots were drawn by the “pheatmap” and “ggplot2”. The R package “forestplot” was used to plot the forest plot. The Kaplan-Meier method was used to assess overall survival curves, and the distinct between survival distributions were assessed with the two-sided log-rank test as implemented in the R package “survival”. A modified drawing survival curve function “gsurvplot”, as implemented in the R package “survminer”, was used for drawing the Kaplan-Meier survival curves in this research. The univariable Cox proportional hazard model was conducted to assess the association between signatures and the patient’s overall survival. All the statistical analyses were performed in R program, and the P-value <0.05 was considered statistically significant.
Construction of the IRPS and the IRPS reflected the immune infiltration status
In order to establish an immune prognostic scoring model related to immune infiltration level, we analyzed 230 immune related signatures. First, we performed ssGSEA on 230 immune related signatures in discovery datasets and calculated the NES for all immune related signatures in each patient. Next, the HRs of the NES in each patient was assessed by univariate Cox regression model, and a meta-analysis was performed on all immune related signatures to assess the overall prognosis. We finally identified 48 immune related signatures with significant prognostic value (HR<1 or HR>1, P<0.05). Among the 48 signatures, the HR values of 31 signatures was less than 1, indicating that the higher the NES, the longer the survival time of patients; However, the HR values of the 17 signatures >1 indicates that the NES is high and the prognosis of patients is poor. Finally, we constructed IRPS based on the NES of 48 immune related signatures (Figure 1 and Table 1).
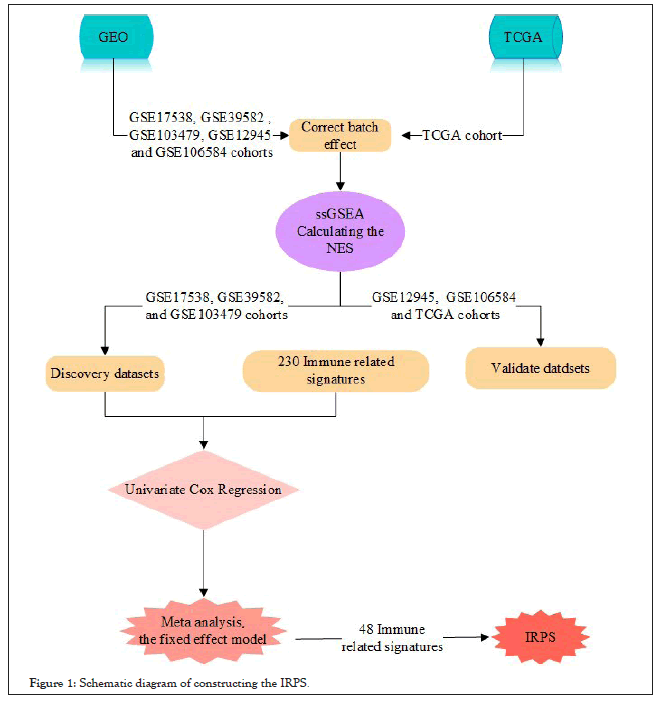
Figure 1: Schematic diagram of constructing the IRPS.
| Signature | HR | CI | P value |
|---|---|---|---|
| EP_BLOOD_VESS_DEVEL_DN_IN_R | 0.3196 | 0.2177-0.4694 | <0.0001 |
| CD103pos_CD103neg_ratio | 0.3589 | 0.2268-0.5679 | <0.0001 |
| WOUNDING_UP_IN_MAPKi_aPDL1_NR | 0.441 | 0.3386-0.5745 | <0.0001 |
| C7_Atom_21 | 0.4603 | 0.3448-0.6145 | <0.0001 |
| Tcm_cells | 0.4644 | 0.3644-0.5919 | <0.0001 |
| MAPKi_INDUCED_ANGIOGENESIS | 0.4861 | 0.3784-0.6244 | <0.0001 |
| Tem_cells | 0.4965 | 0.3794-0.6497 | <0.0001 |
| Mast_cells | 0.4993 | 0.3819-0.6527 | <0.0001 |
| Th1_cells | 0.5035 | 0.3817-0.6642 | <0.0001 |
| HYPOXIA_UP_IN_MAPKi_aPDL1_NR | 0.5056 | 0.3759-0.6802 | <0.0001 |
| TGFβ_pathway | 0.5099 | 0.3762-0.6909 | <0.0001 |
| DTPP_REG_CELL_PROLIF_UP | 0.5108 | 0.3959-0.6591 | <0.0001 |
| Core_Inhibition | 0.5203 | 0.4194-0.6455 | <0.0001 |
| DTP_BLOOD_VESS_DEVEL_UP | 0.5261 | 0.4094-0.6761 | <0.0001 |
| CD68 | 0.5409 | 0.4319-0.6775 | <0.0001 |
| Immunosupppressive_core_pathways | 0.5543 | 0.4433-0.6930 | <0.0001 |
| MCD3_CD8 | 0.5631 | 0.4490-0.7062 | <0.0001 |
| Danger_signal_pathways | 0.5644 | 0.4341-0.7337 | < 0.0001 |
| GRANS_PCA | 0.5665 | 0.4443-0.7223 | < 0.0001 |
| PH_RESP_TO_WOUNDING_DN_IN_R | 0.5756 | 0.4656-0.7116 | <0.0001 |
| PH_BLOOD_VESS_DEVEL_DN_IN_R | 0.5864 | 0.4630-0.7427 | <0.0001 |
| C7_Atom_10 | 0.5876 | 0.4554-0.7582 | <0.0001 |
| PLX2D_CELL_ADHESION_UP | 0.5914 | 0.4770-0.7332 | <0.0001 |
| TGFB_PCA | 0.5976 | 0.4743-0.7530 | <0.0001 |
| C7_Atom_22 | 0.602 | 0.4719-0.7679 | <0.0001 |
| C7_Atom_11 | 0.6041 | 0.4804-0.7595 | <0.0001 |
| MAPKR_BLOOD_VESS_DEVEL_UP | 0.6042 | 0.4679-0.7802 | 0.0001 |
| NK_cells | 0.6214 | 0.4978-0.7756 | <0.0001 |
| HIPPO_pathway | 0.6256 | 0.5002-0.7825 | <0.0001 |
| Core_Activation | 0.6324 | 0.4887-0.8183 | 0.0005 |
| G_CYTH4 | 0.6354 | 0.5036-0.8017 | 0.0001 |
| CD8A | 1.2367 | 1.0992-1.5306 | 0.0051 |
| NK_cell | 1.2543 | 1.0120-1.5547 | 0.0386 |
| T_cells | 1.5277 | 1.2134-1.9234 | 0.0003 |
| CSR_Activated | 1.6003 | 1.2637-2.0267 | <0.0001 |
| NRF2_pathway | 1.6136 | 1.2972-2.0071 | <0.0001 |
| G_CD3E | 1.6849 | 1.3133-2.1617 | <0.0001 |
| T_cell | 1.6982 | 1.2935-2.2295 | 0.0001 |
| Tcell_receptors | 1.7123 | 1.3707-2.1389 | <0.0001 |
| C7_Atom_15 | 1.7735 | 1.3691-2.2975 | <0.0001 |
| Module5_TcellBcell | 1.8306 | 1.4016-2.3910 | <0.0001 |
| TIS_signature | 1.8336 | 1.4199-2.3679 | <0.0001 |
| TAMsurr_TcClassII_ratio | 1.8961 | 1.4248-2.5232 | <0.0001 |
| T_helper_cells | 1.9174 | 1.4740-2.4941 | <0.0001 |
| APMS | 2.0022 | 1.5968-2.5105 | <0.0001 |
| IL8 | 2.2732 | 1.7192-3.0057 | <0.0001 |
| ICR | 2.3475 | 1.8145-3.0370 | <0.0001 |
| ICR_ACT | 2.4897 | 1.9175-3.2326 | <0.0001 |
Table 1: Immune related signature.
Among the 48 immune related signatures, the infiltration levels of immune cells (T cells, Th1 cells, Tcm cells, Tem cells, NK cells and mast cells) plays a role for the prognosis of patients as shown in Figure 2A. It is well known that T cells and NK cells play an immune promoting role in the anti-tumor process [7-9]. We observed that the IRPS was significantly negatively related to immune promoting cells, such as T cells and NK cells; however, the IRPS showed positive correlation with immunosuppressive cells (Th1 cells, Tcm cells, Tem cells and mast cells) as shown in Figures 2B and 2C. We observed similar results in the validation dataset. These results show that the IRPS may play an immunosuppressive role in the anti-tumor process.
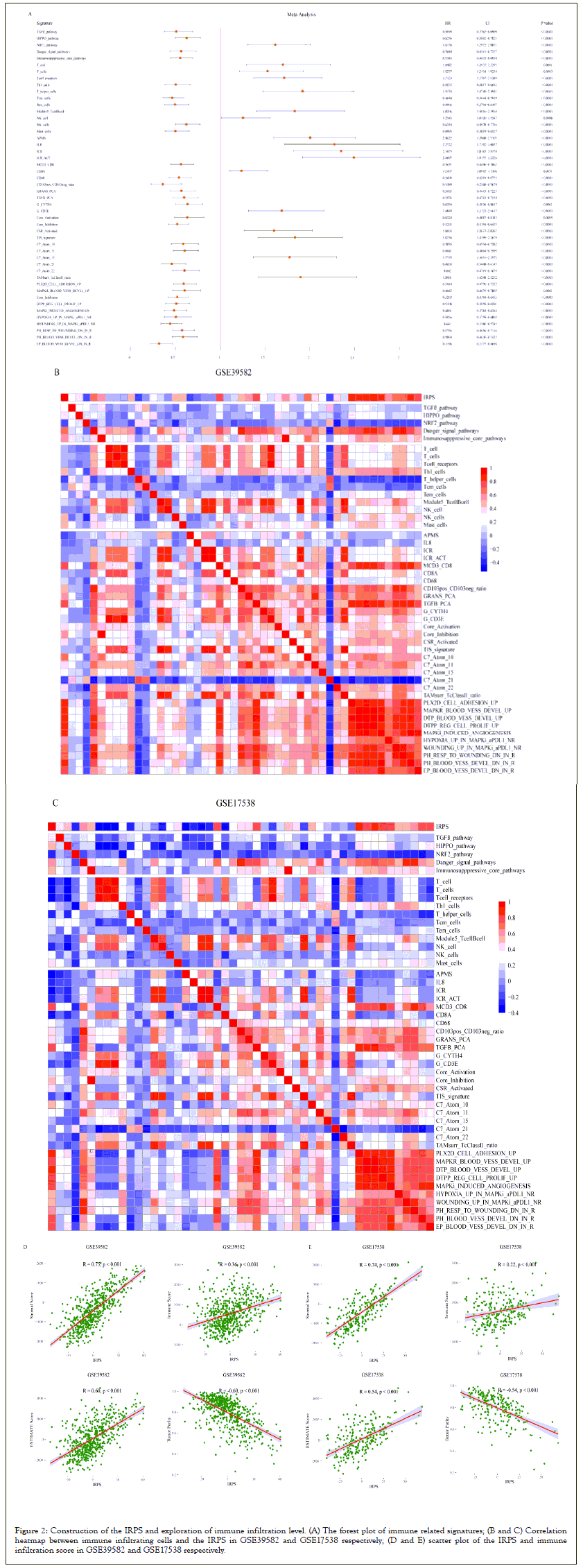
Figure 2: Construction of the IRPS and exploration of immune infiltration level. (A) The forest plot of immune related signatures; (B and C) Correlation heatmap between immune infiltrating cells and the IRPS in GSE39582 and GSE17538 respectively; (D and E) scatter plot of the IRPS and immune infiltration score in GSE39582 and GSE17538 respectively.
In addition, the level of immune infiltration also plays an important role in the anti-tumor process. Therefore, we calculated the immune infiltration score for each sample, including ESTIMATE score, immune score, stromal score, and tumor purity. By analyzing the correlation between the IRPS and immune infiltration score, we observed that the IRPS was significantly positively correlated with ESTIMATE score, immune score and stromal score. On the contrary, the IRPS was negatively correlated with tumor purity as shown in Figures 2D and 2E.
We observed that the IRPS constructed by the NES according to immune related signatures have immune inhibiting effects in the process of anti-tumor immunity and can reflect the level of immune infiltration in CRC patients (Figures 2A-2E).
Relationship between the IRPS and microsatellite status in colorectal cancer
We divided all patients into the high IRPS group and the low IRPS group according to the best cut-off of the IRPS. Subsequently, we compared the overall survival of patients in different the IRPS groups. We found that there was a significant difference in overall survival between the high IRPS group and the low IRPS group by survival analysis of the discovery datasets. In GSE39582 and GSE17538 cohorts, the overall survival time of patients in the low IRPS group was significantly longer than that in the high IRPS group as shown in Figure 3A; however, in the GSE103479 cohort, we observed the opposite result, although the analysis results were not statistically significant. Meanwhile, we also carried out survival analysis on the validation datasets, we observed the similar result in the three validate cohorts, that is, the overall survival time of patients in the low IRPS group was significantly longer. This suggests that the IRPS may act as a risk factor in CRC patients.
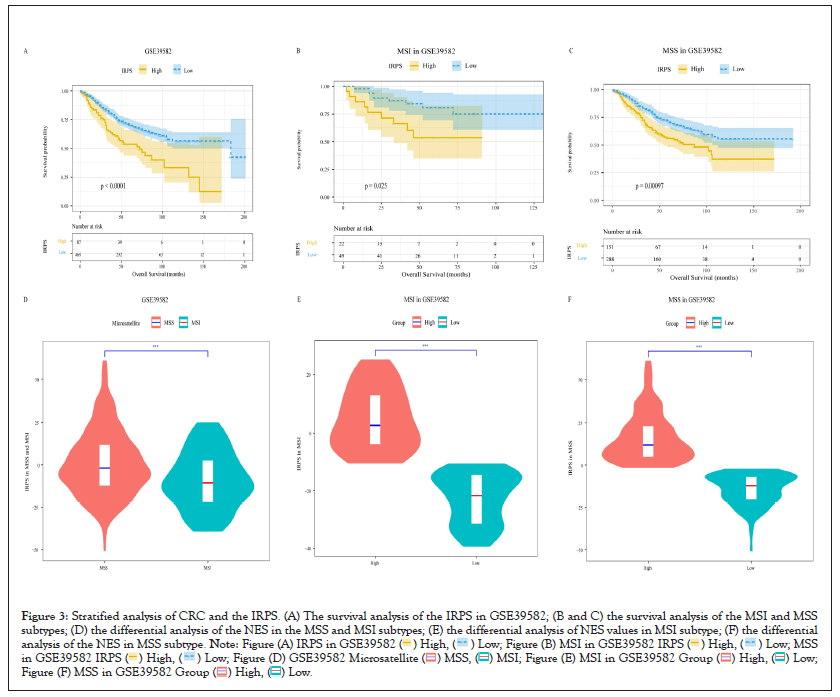
Figure 3: Stratified analysis of CRC and the IRPS. (A) The survival analysis of the IRPS in GSE39582; (B and C) the survival analysis of the MSI and MSS
subtypes; (D) the differential analysis of the NES in the MSS and MSI subtypes; (E) the differential analysis of NES values in MSI subtype; (F) the differential
analysis of the NES in MSS subtype. 
We know that the microsatellite status of CRC patients is an important factor affecting their prognosis and affects the patient's response to immunotherapy. To further clarify the impact of the IRPS on CRC patients, we performed an association analysis between the IRPS and microsatellite status. In our cohorts, only the GSE39582 and TCGA cohorts recorded microsatellite status information of patients, so we only further analyzed these two cohorts. Classifying CRC patients into Medical Student Section (MSS) subtype and Microsatellite Instability (MSI) subtype according to the microsatellite status of each patient. We first performed survival analysis on patients in different subtypes. This is obvious, in both the discovery and validation dataset, patients in the low IRPS group had a longer survival time as show in Figures 3B and 3C. In the TCGA cohort, the survival analysis results of MSI subtype patients were not statistically significant, which may be due to the small number of samples. Then, we found the IRPS is significantly lower in patients with MSI subtype as show in Figure 3D. We further analyzed the distinction between the high IRPS and the low IRPS in the different microsatellite status. We can view that there is significant difference between the high IRPS group and the low IRPS group in two microsatellite status subtypes, and the IRPS is significantly lower in patients with MSI subtype as shown in Figures 3E and 3F.
These results prove that the IRPS is a prognostic factor of CRC. At the same time, by analyzing the differences of the IRPS in patients with different microsatellite status, we find that the level of the IRPS may potentially reflect the patient's response to immunotherapy in different microsatellite status (Figures 3A-3F).
Association between innate immune escape mechanism and the IRPS in colorectal cancer
The intrinsic immune escape mechanism of tumor to immunotherapy mainly has two aspects, immunogenicity and immune checkpoint molecular expression [26]. Several potential prognostic factors, including TMB, Homologous Recombination Deficient (HRD), neo-antigen load, Loss Of Heterozygosity (LOH), Copy Number Variation (CNV) and Single Nucleotide Variation (SNV), determined tumor immunogenicity. In order to explore the effect of the IRPS on innate immune escape of CRC, we analyzed the relationship between the IRPS and tumor immunogenicity of CRC. In all our cohorts, the validation dataset TCGA cohort has complete genomic information, so we used the TCGA cohort for further analysis.
By comparing the differences of these potential prognosis factors between the high IRPS group and the low IRPS group, we found the prognostic factors CNV, LOH and HRD were higher in the low IRPS subtype than in the high IRPS subtype as shown in Figures 4A-4H. Opposite, the median values of prognostic factors immunogenic indel, indel, TMB, neo-antigen load and SNV tend to be highly expressed in the high IRPS subtype as shown in Figures 4B-4G. We further analyzed the correlation between the IRPS and prognostic factors. Unsurprisingly, the IRPS was negatively correlated with CNV, LOH and HRD as shown in Figures 5A-5H. Simultaneously, the IRPS was positively correlated with immunogenic indel, indel, TMB, neoantigen load and SNV as shown in Figures 5B-5G.
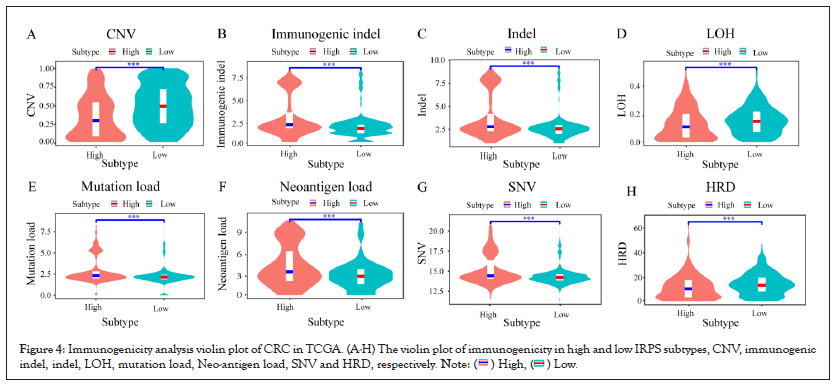
Figure 4: Immunogenicity analysis violin plot of CRC in TCGA. (A-H) The violin plot of immunogenicity in high and low IRPS subtypes, CNV, immunogenic
indel, indel, LOH, mutation load, Neo-antigen load, SNV and HRD, respectively. 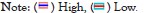
Figure 5: Immunogenicity analysis scatter plot of CRC in TCGA. (A-H) The violin plot of immunogenicity in the high and low IRPS subtypes, CNV, immunogenic indel, indel, LOH, mutation load, Neo-antigen load, SNV and HRD, respectively.
As a prognostic factor of MSI patients, MSI patients with high TMB are effective for immunotherapy. Given the significant differential expression of the IRPS in MSI tumors as shown in Figure 3E and the positive correlation between the IRPS and TMB as shown in Figure 4E, we speculated that genomic changes in tumors might remodel the immune phenotype (Figures 4A- 4H and Figures 5A-5G).
Exploring relationship between the IRPS and genomic alterations in colorectal cancer
To verify whether the immune phenotype is remodeled by genomic alterations of the tumor, we analyzed the associations between the IRPS and somatic mutations in the TCGA cohort. Firstly, we calculated the somatic mutation frequency of each gene in the cohort and visualized the somatic mutation frequency of the top 35 genes. We observed that the genes with the highest somatic mutation frequency in CRC were Antigen-Presenting Cell (APC) and TP53, followed by Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) and Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA). These genes with high mutation frequency are common in some carcinogenic pathways, including cell cycle pathway, Hippo signaling, MYC signaling, Neurogenic Locus Notch Homolog Protein (NOTCH) signaling, oxidative stress response / Nuclear Factor Erythroid 2 (NRF2), PI-3-Kinase signaling, Receptor-Tyrosine Kinase (RTK) / Renin-Angiotensin-System (RAS) / Mitogen-Activated Protein Kinase (MAPK) signaling, Transforming Growth Factor (TGF-β) signaling, TP53 pathway and Wingless-Related Integration Site (WNT) /β-catenin signaling21.
Subsequently, we discussed the difference of NES of 10 oncogenic pathways between the high IRPS and the low IRPS subtypes in TCGA cohort as shown in Figures 6A-6L. It is obvious that the NES of most of the 10 carcinogenic pathways are higher in low IRPS subtypes, in particular, Hippo signaling, MYC signaling, oxidative stress response/NRF2, PI-3-Kinase signaling and TP53 pathway. Only the NES of cell cycle pathway and NOTCH signaling is higher in the high IRPS subtype. This seems to verify that somatic mutations in common carcinogenic pathways can affect the immuno-phenotype of patients with CRC (Figures 6A-6L).
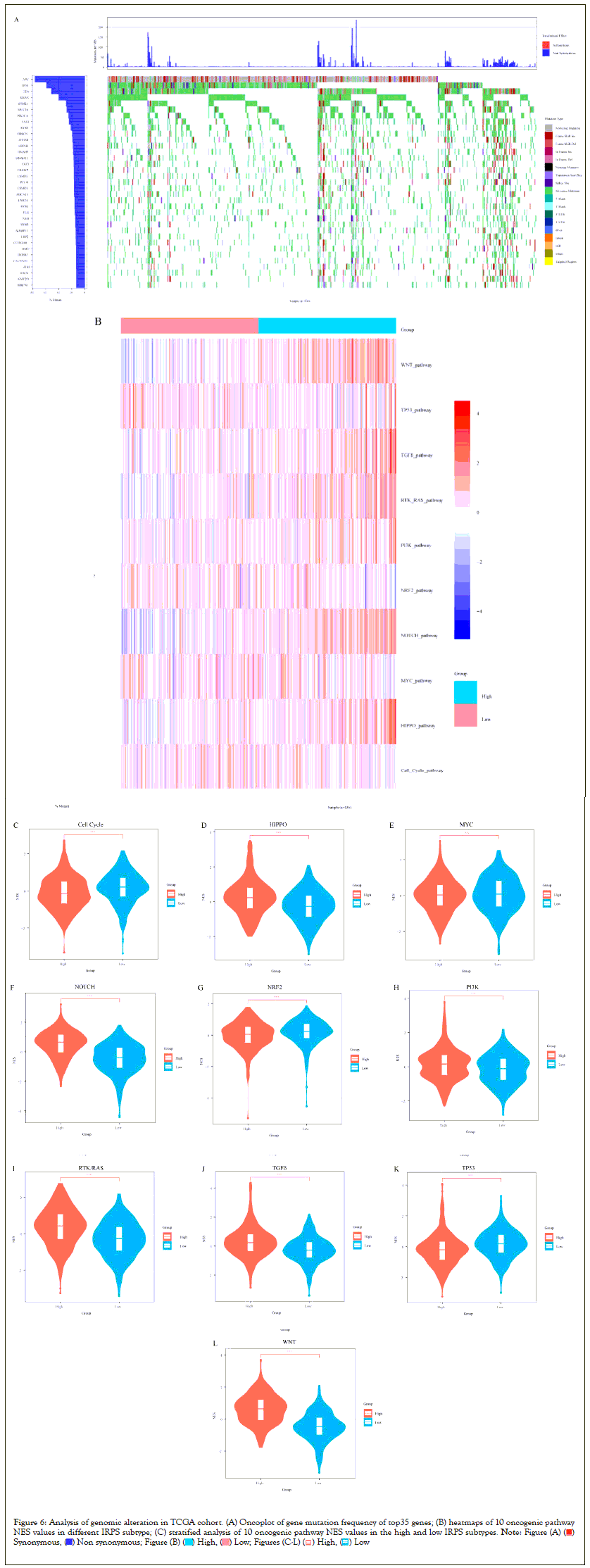
Figure 6: Analysis of genomic alteration in TCGA cohort. (A) Oncoplot of gene mutation frequency of top35 genes; (B) heatmaps of 10 oncogenic pathway
NES values in different IRPS subtype; (C) stratified analysis of 10 oncogenic pathway NES values in the high and low IRPS subtypes. 
The IRPS could predict the immunotherapeutic benefit
Based on the results of the previous analysis, it is suggested that there may be a potential association between the IRPS and immunotherapy of CRC, so we further analyzed the immunotherapeutic benefits of the IRPS in CRC. For doing this, we need to obtain the datasets of CRC immunotherapy firstly, but after searching the published CRC immunotherapy datasets, there is no suitable cohorts related to CRC immunotherapy. Instead, we used melanoma and bladder cancer immunotherapy cohort (GSE78220 and IMvigor210) to test the immunotherapeutic benefits of the IRPS.
Taking the best cut-off value of the IRPS as the cut-off value, patients in GSE78220 and IMvigor210 cohorts were divided into the high IRPS subtype and the low IRPS subtype. Patients in the low IRPS subtype had longer overall survival compared with patients in the higher IRPS subtype in the GSE78220 cohort (P-value=0.0079) as shown in Figures 7A-7J and IMvigor210 cohort (P-value=0.0012) as shown in Figure 7E. The violin diagram showed that the IRPS of patients who responded to immunotherapy decreased significantly compared with patients who did not respond to immunotherapy as shown in Figures 7B and 7F. The association between immunotherapy response and existing immune activity was analyzed by using the information about immunotherapy response in two immunotherapy cohorts we collected. We observed that patients in the low IRPS subtype responded better to immunotherapy than patients in the higher IRPS subtype as shown in Figures 7C and 7G. Finally, the waterfall plot of two immunotherapy datasets illustrates more patients with CR/PR for the immunotherapy have lower the IRPS value than PD/SD patients for the immunotherapy as shown in Figures 7D and 7H.
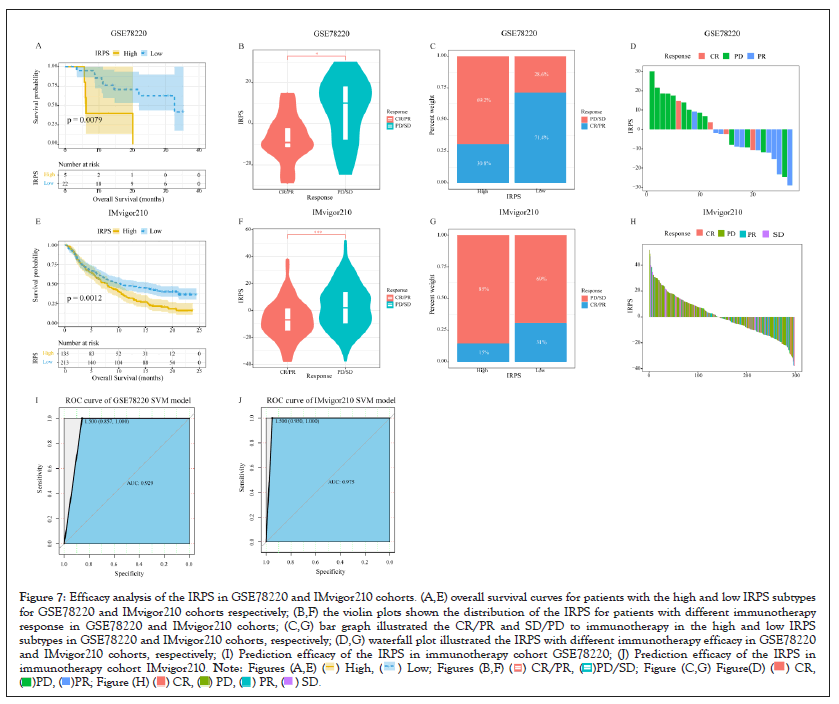
Figure 7: Efficacy analysis of the IRPS in GSE78220 and IMvigor210 cohorts. (A,E) overall survival curves for patients with the high and low IRPS subtypes
for GSE78220 and IMvigor210 cohorts respectively; (B,F) the violin plots shown the distribution of the IRPS for patients with different immunotherapy
response in GSE78220 and IMvigor210 cohorts; (C,G) bar graph illustrated the CR/PR and SD/PD to immunotherapy in the high and low IRPS
subtypes in GSE78220 and IMvigor210 cohorts, respectively; (D,G) waterfall plot illustrated the IRPS with different immunotherapy efficacy in GSE78220
and IMvigor210 cohorts, respectively; (I) Prediction efficacy of the IRPS in immunotherapy cohort GSE78220; (J) Prediction efficacy of the IRPS in
immunotherapy cohort IMvigor210. 
In the cohort of GSE78220 and IMvigor210, the IRPS significantly related to the efficacy of immunotherapy was evaluated by predictive model. By using the IRPS as input parameter of Support Vector Machine (SVM), support vector machine is implemented in R software package “e1071”. Radial Basis Function (RBF) is selected as the kernel function of support vector machine. The grid search is used to optimize the penalty parameter C and kernel coefficient gamma of support vector machine. The IRPS subtype of patients is used as the prediction target of the classifier. In the test, the overall accuracy of GSE78220 and IMvigor210 cohort in predicting patients' immunotherapy response was 0.929 and 0.975, respectively as show in Figures 7I and 7J. The predictive power of the two immunotherapy cohorts clearly shows that the IRPS is a predictive biomarker of immunotherapy benefits (Figures 7A-7J).
In this research, we included more than 1800 CRC patients in 6 cohorts for analysis. We used ssGSEA to calculate the NES of each patient, separately. The ssGSEA method does not rely on the influence of other samples in the gene expression datasets when scoring a single sample, and provides stable scores that are unlikely to be affected by different sample and gene sizes in the cohorts and unnecessary changes between samples. Thus, the ssGSEA score of each patient is less affected by the microarray platform of each cohort. The univariate Cox regression model results of 18 cohorts were analyzed by meta-analysis, and screening 48 immune related signatures. Finally, the IRPS of each patient is calculated through the NES of 48 signatures. Specifically, for each patient, we calculated the sum of the NES values of signatures with HRs less than 1 and HRs more than 1 respectively, and then the IRPS is equal to the subtraction of them.
In the 6 CRC datasets collected, most patients with the low IRPS showed better overall survival than those with the high IRPS. The results show that the IRPS can reflect the level of immune infiltration in the tumour microenvironment of CRC patients. The IRPS is negatively related to anti-tumour immune infiltrating cells and positively related to tumour immunosuppressive cells. It is well known that immunotherapy of CRC patients is closely related to microsatellite status and immune checkpoint molecules [11,14,27-29]. We confirmed the correlation between the IRPS and immunotherapy by analysing the relationship between the IRPS and microsatellite status. In addition, the potential mechanism of the IRPS affecting immunotherapy was explored based on genomic analysis. Among the many immunogenicity factors that affect the response to immunotherapy, TMB is a major factor, and TMB is related to the IRPS. At the same time, the relationship between the IRPS and other immunogenicity also further indicates that the IRPS may affect the response of immunotherapy by affecting immunogenicity [30].
In recent years, ICIs, such as anti-PD-1, anti-PD-L1 or anti-CTLA-4 therapy, in patients with solid tumours has aroused great interest in immunotherapy. Nevertheless, only a few patients have a long and stable response during immune checkpoint inhibiters treatment. Lately, TMB and the expression level of PD-1 and PD-L1 have been found not to be effective biomarkers to predict the benefits of immune checkpoint blocking [31]. Therefore, it is necessary to develop a useful tool to predict the efficacy of immunotherapy. In this study, the predictive ability of the IRPS for checkpoint inhibitors was demonstrated in two groups of advanced melanoma and bladder cancer patients treated with anti-PD-1 antibody and anti-CTLA4 antibody, respectively. Our results distinctly show that the IRPS is associated with the response to different immunotherapy methods, including immunotherapy based on anti-PD-1, anti-PD-L1 and CTLA4 immune checkpoint inhibitors. The prediction accuracy of the two groups of patients indicated that the IRPS value can be used as a tool to evaluate the effectiveness of immunotherapy for cancer. The IRPS of patients who responded to immunotherapy were significantly lower than that of patients who did not respond to immunotherapy. Owing to the lack of a CRC cohort receiving immunotherapy, more prospective trials are needed to further validate these findings in patients with CRC.
There are still some deficiencies in this study. First, we obtained as many CRC cohorts as possible, but still did not find a suitable CRC immunotherapy cohort. We also need to conduct more experimental studies on immune related signatures, in order to provide more available information for further understanding their function in CRC. Finally, we only use the best cut-off of the IRPS to classify CRC samples into high IRPS subtype and low IRPS subtype, however, no other grouping method is used. We still need to find better cut-off values to provide a better strategy for stratification of CRC patients.
The IRPS is a valuable signature related to the level of immune infiltration. The correlation between the IRPS and the overall survival rate of CRC comprehensive cohort proves that the IRPS is a powerful prognostic hallmark of CRC. The IRPS should contribute to further determining the prognosis of patients with CRC and predict the response of patients with CRC to immunotherapy.
The IRPS is a valuable signature related to the level of immune infiltration. The correlation between the IRPS and the overall survival rate of CRC comprehensive cohort proves that the IRPS is a powerful prognostic hallmark of CRC. The IRPS should contribute to further determining the prognosis of patients with CRC and predict the response of patients with CRC to immunotherapy.
The authors would like to thank TCGA, PubMed and GEO projects for the data access. Thanks to Research Square for allowing us to publish a preprint of the manuscript.
This work was supported by the National Nature Science Foundation of China (grant number 62276084), Central support for the reform and development of local colleges and universities (high-level talent project) (grant number 2020GSP05), Heilongjiang Province Applied Technology Research and Development Plan Project (GA19C003) and the National Natural Science Foundation of China Youth Science Fund Project (grant number 82103030).
The authors have no relevant financial or non-financial interests to disclose.
The datasets analysed during the current study are available in the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) repository. The web link of GEO is https://www.ncbi.nlm.nih.gov/geo/, datasets in GEO includes GSE12945, GSE17538, GSE39582, GSE103479 and GSE106584. The web link of TCGA is https://portal.gdc.cancer.gov/repository.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Jun Xiang, JunHu Li and XiaoNan Cheng. The first draft of the manuscript was written by Jun Xiang and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
The authors confirm that there are no conflicts of interest.
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Harbin Medical University.
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar][PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
Citation: Xiang J, Li J, Cheng X, Hu H, Yu L, Wang G (2023) Constructing the Immune Related Prognosis Score to Predict the Benefit of Immunotherapy in Colorectal Cancer. Immunogenet Open Access.8:199.
Received: 24-Mar-2023, Manuscript No. IGOA-23-22341; Editor assigned: 28-Mar-2023, Pre QC No. IGOA-23-22341 (PQ); Reviewed: 11-Apr-2023, QC No. IGOA-23-22341; Revised: 18-Apr-2023, Manuscript No. IGOA-23-22341 (R); Published: 25-Apr-2023 , DOI: 10.35248/IGOA.23.8.199
Copyright: © 2023 Xiang J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : no