Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research - (2020)Volume 9, Issue 1
Multi-Alignment method coupled with phylogenetic analysis we disclosed the Nsp9 and Nsp10 non-structural proteins of Corona Virus asrRNARlmH/K methyltransferases with similarities with bin recombinase and int-core integrase fold. Further, Nsp9 has similarities to S8 ribosomal protein and Nap10 has similarity to S10 ribosomal protein. Previously, we showed Nsp13, Nsp14, Nsp15 and Nsp16 are also different types of rRNARlmE/N and Cfr-like methyltransferases-ribonucleasewith RNA helicase domains. Two domains of Nsp13 astonishingly have similarities to ribosomal proteins L6 and L9. Taken together, Nsp9/10 and Nsp13-16 proteins could mimic host ribosome assembly and also could methylate rRNA of mitobibosome preventing mitochondrial protein synthesis and oxidative phosphorylation. Low ATP synthesis causes lowering blood pressure following coma but very ATP concentration (1-10nM) surely induces platelets aggregation through vWA, collagen and GpIIb/IIIaproteins followed byfibrin formation and blood clotting as recently have seen in the lung of many Corona virus infected patients. We have also postulated that two polyproteins itself resemble like 28S and 38S mitoribosome subunits and compete with rRNAs inhibiting the ribosome turnover and new protein synthesis due to their similarities with many ribosomal proteins.Such finding may be valuable in computer-based novel drug design against Corona virus.
Coronavirus Nsp proteins;Ribosomal proteins homology; Inhibition of ribosome turn over; rRNAmethyltransferase; Protein synthesis inhibition;Low ATP formation; Blood clotting; Low blood pressure and coma
Coronaviruses (family Coronaviridae) are enveloped viruses with a largest positive sense, single-stranded RNA genome of ~30kb [1,2]. On genetic and antigenic criteria, CoVs have been organised into three different groups: α-CoVs, β-CoVs, and γ-CoVs [3-5]. Coronaviruses primarily infect birds, mammals and human, causing many lethal respiratory syndromes resembling the common cold, such as bronchitis, pneumonia, and even severe acute respiratory syndrome (SARS). Recently corona viral research augmented due to pandemic severe respiratory illnesses outbreaks claiming >100000 deaths due to human Corona virus [6]. COVID-19 virus enters cells through ACE2 receptor-mediated endocytosis in lungs alveolar epithelial cell as well as cells in the heart and kidney [7].
The extremely large gene 1 covers 2/3 of the 30 kb genome (separated into ORF1a and orf1b) encoding polyproteins which proteolytically degraded into sixteen non-structural proteins involved in mRNA synthesis and replication of virus through (-) strand RNA synthesis by RNA-dependent RNA polymerase. The rest 1/3 of the 3’end of the RNA genome encodes the structural spike glycoprotein (S), small envelope protein (E), membrane glycoprotein (M), and nucleocapsid protein (N) as well as few small transcripts like ORF2b, ORF7a and ORD2a (see, accession no. DQ415908, KT779555, KY674941, AY884001, DQ415912, KY674941, DQ415914, MH940245).
The function of Nsp2 was proposed by me as RNA topoisomerase and Nsp13 was determined as 2’-O-ribose capping Guanosinemethyl transferase. By bioinformatics approach comparing 200 DNA and MTases, Ligases, RNases, DNases, Ribosomal proteins as well as some RNA virus associated non-structural proteins we wanted to determine the functions of nsp9 and Nsp10 of Corona virus [8,9]. We proposed Nsp16 as RlmE-type 2’-O-ribose Uridine methyl transferase and Nsp14 as N7 Guanidine methyl transferase whereas now we proposed Nsp9 as RlmG-like and Nsp10 as ErmD-type methyl transferase accounting total five rRNA methyl transferases in Corona virus genome.
The rRNAMTasesmethylate at least nine 23S rRNA nucleotides (G748, A1067, C1920, A2058, G2445, G2470, U2479, A2503, T2504 and G2535) on the large ribosomal subunit [10]. There are more than ten 16S rRNA modifying MTases (ArmA, RmtA to RmtH and NpmA) have characterized where as ArmA and RmtH are abundant. Ribosome decoding centre (nucleotides 1400–1500 of 16S rRNA) is the binding sites for aminoglycosides. Endogenous methyl transferases RsmI and RsmH methylate C1402 whereas RsmEmethylatesU1498, and RsmFmethylates C1407 (Tscheme et al.). Different Rlmmethyl transferases methylate at various positions of bacterial 23S rRNA conferring multi-resistant to macrolides and ketolides. As for example, RlmAIIMTase has preference to N1 of G748 of 23S rRNA [11], RlmBMTase (protein id. BAI33654) modifies G2251 [12,13] while RlmC modifies m5U747 and RlmD is specific for m5U1939. RlmE and RlmF methylate 23S rRNA 2’- O-U2552 while methylates at N2 of G1835 and N3 pseudo-Uridine forRlmHand RlmNmethylates C2 at A2503 [14]. RlmM (YgdE; EC:2.1.1.186) enzyme catalyzes the SAM-dependent 2' O-ribose methylation of C2498 in 23S rRNA of Escherichia coli [15]. DcmMethyltransferase (EC:2.1.1.137) causes DNA methylation at the C5 or N4 positions of cytosine. E. coli Dam methyltransferase has GATC sequence specificity and methylates at the adenine residue at N6 regulating many genes [16].
We investigated the homology profiles of many recombinases and transposases using CLUSTAL-Omega software. Several thousand IS elements were sequenced but could be classified into ∼20 families (IS2, IS30, Tn10, Tn21 etc) on the basis of the sequences of their transposases and terminal inverted repeats as well as associated antibiotic resistant, drug efflux and metal resistant genes as found in integrons and transposons [17-19]. Int1I, int and Rci are different integrases [20]. Escherichia coli RecA, RecB and recC have roles in homologous recombination, DNA repair, and the induction of the SOS response [21,22]. Bacterial and mammalian RNases are also RNA modifying enzymes and exonucleasesare also compared [23-25]. We also compared sixty 30S plus 50S ribosomal proteins as they are powerful RNA binding proteins (Hammerling et al.). We have compared amino acid sequences of those 100-150 proteins with Nsp9 and Nsp10 non-structural proteins related to polyprotein ORF1ab of coronavirus. BLAST search and Multi-alignment analysis are important tools to find the function of Unknown viral protein like Nsp9/10 of Coronavirus. Previously, Nsp2 and Nsp13 functions were predicted as RNA helicase-topoisomerase but and capping Guanine 2’-O-Ribose methyltransferase respectively [26,27].
The BLAST search was done using web portal www.ncbi.nlm. nih.gov/blast and retrieve of covid-19 and other Corona viruses cDNA sequences were done using web portal www.ncbi.nlm.nih. gov/nucleotide or protein. NCBI Primer Design Software was used for primer selection and Oligoanalyzer 3.2 software was used to analyze primer dimmer and hairpin structure. Multalin Software and CLUSTAL Omega Software were used to multiple align of protein sequences and NCBI BLAST seq-2 analysis portal used to analyze homology between two sequences. NCBI PubMed portal (www.ncbi.nlm.nih.gov/pubmed) used to retrieve references and papers. CLUSTAL Omega Phylogenetic tool used to determine the closer structural similarities among the proteins and Seq-2 BLAST was confirmed percentage of sequence homology between two related proteins [28,29].The structure and localization of Corona proteins were demonstrated in Figure 1.
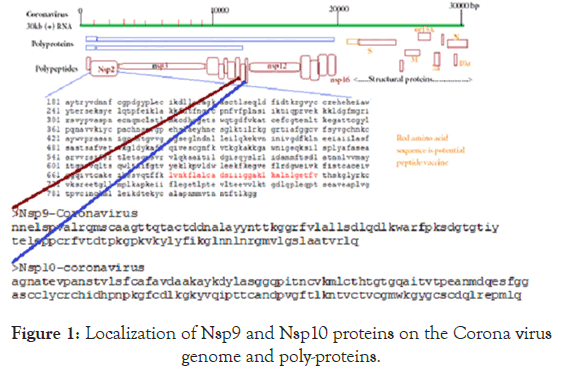
Figure 1. Localization of Nsp9 and Nsp10 proteins on the Corona virus genome and poly-proteins.
BLAST and CLUSTAL Omega software’s are very powerful to compare unknown DNA and protein molecules as vast number of genes were deposited in the GenBank and crystal structures and functional domains of protein sequences were known. Figure 2 disclosed the multialign-phylogenetic relations of 50 DNA/RNA modifying genes with Nsp9 and Nsp10 non-structural proteins of Corona virus and the functions of such proteins are still elusive. It was clearly indicated that Nsp9 share a relation with ermD and Nsp10 with exonuclease or RlmK. Figure 3 showed the multi-alignment and phylogenetic relation of Nsp9 and Nsp10 with seventy five Escherichia coli ribosomal proteins.It is clearly indicated that Nsp9 has close similarity to S8 and Nsp10 to S10 and L22 ribosomal proteins.

Figure 2. Multialignment of different DNA/RNA modifying enzymes with Nsp9/10 of Corona virus.

Figure 3. Multialignment of 30S and 50S ribosomal RNA binding proteins with Nsp9/10 of Corona virus.
We detected about 20%-25% scattered homologies between Nsp9 and S8 ribosomal protein (Figure 4A) and between Nsp10 and S10 ribosomal protein (Figure 4B). In Figure 5 we demonstrated the similarities among ErmDmrthyltransferase with Nsp9 and RlmGmethyl transferase, int-core integrase and bin recombinase with Nsp10 protein of Corona virus. (A) Nsp9 vs. ErmD, (B) Nsp10 vs. RlmG, (C) was Nsp10 vs. bin-recombinase and (D) Nsp10 vs. int-core integrase. The ErmD has also similarities to RmtH and intintegrase but such similarities are not great and expected for distance search between viruses vs, Bacteria. The result might be confusing but problem solved when we did homology search between RlmG vs. bin-recombinase and RlmGvs. int-core integrase (Figure 6) showing homology profile of those enzymes. The problem is BLAST multi-alignment hardly able to catch such similarities and only gave maximum homology at certain point of the sequence. Similarly, CLUSTAL multi-alignment search drastically made gaps and hard to interpret homology data unless we made phylogenetic conversion.
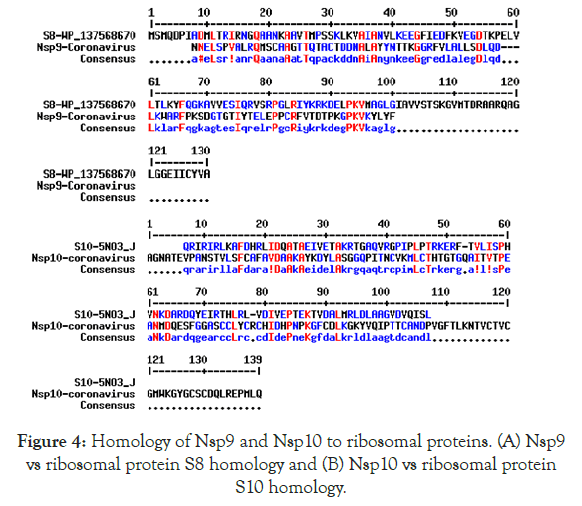
Figure 4. Homology of Nsp9 and Nsp10 to ribosomal proteins. (A) Nsp9 vs ribosomal protein S8 homology and (B) Nsp10 vs ribosomal protein S10 homology.
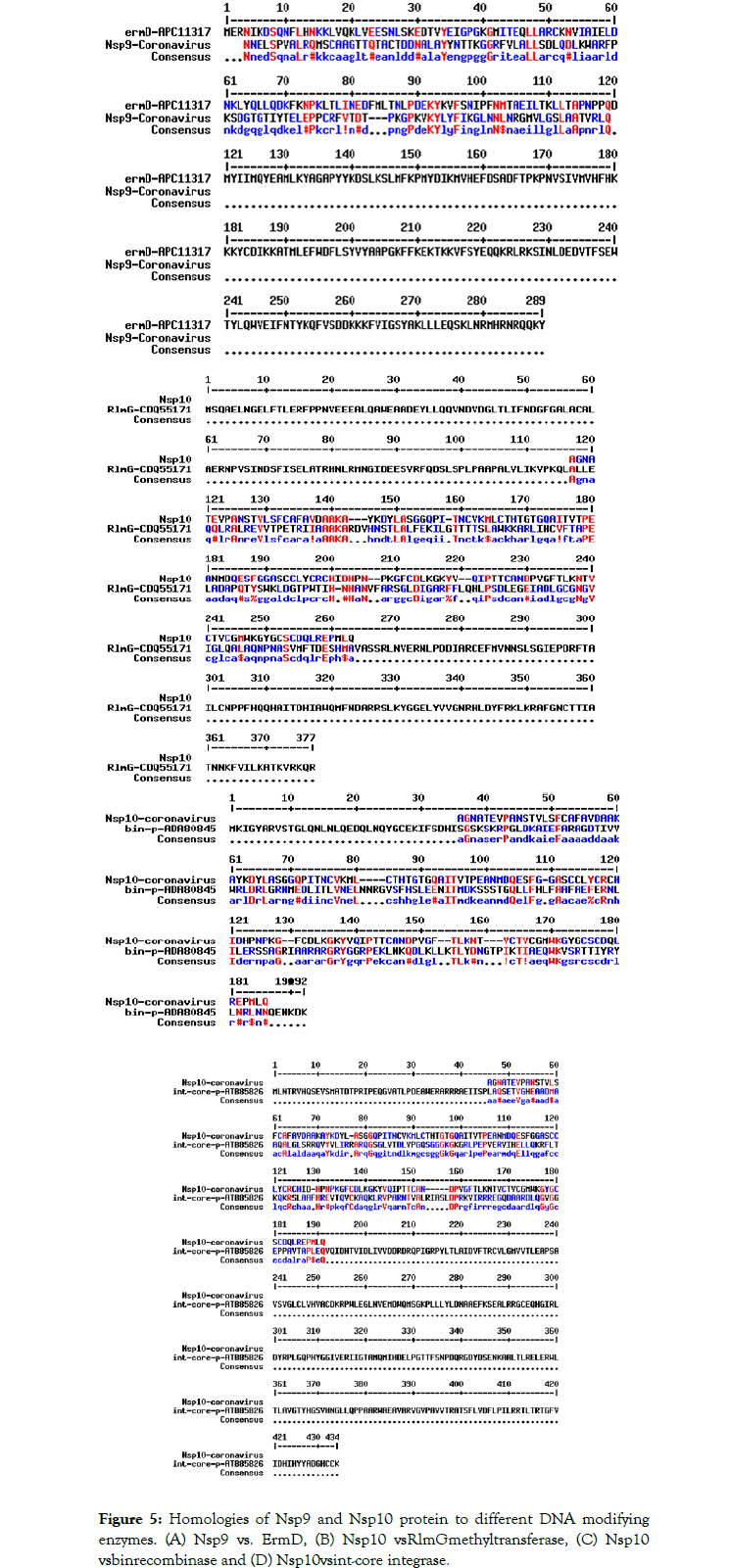
Figure 5. Homologies of Nsp9 and Nsp10 protein to different DNA modifying enzymes. (A) Nsp9 vs. ErmD, (B) Nsp10 vsRlmGmethyltransferase, (C) Nsp10 vsbinrecombinase and (D) Nsp10 vsint-core integrase.
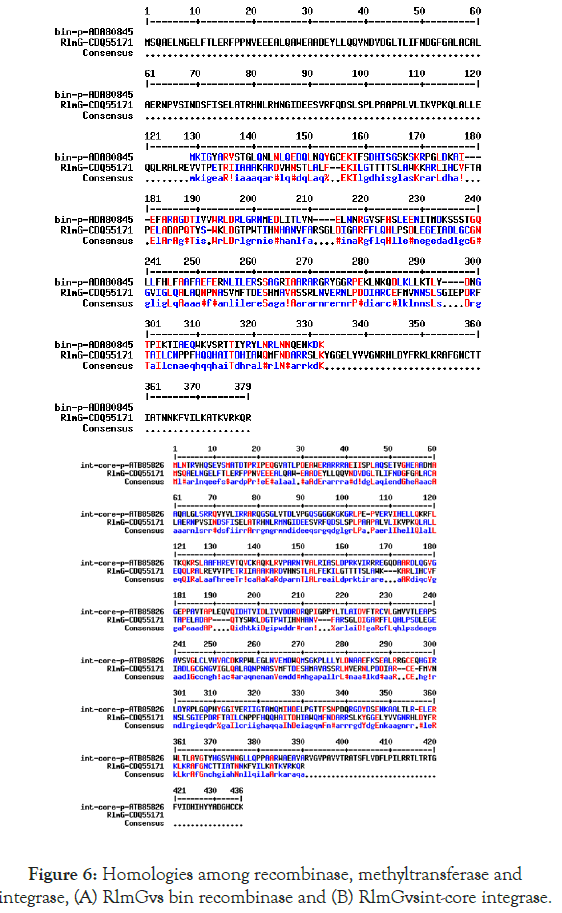
Figure 6. Homologies among recombinase, methyltransferase and integrase, (A) RlmGvs bin recombinase and (B) RlmGvsint-core integrase.
Previously, Nsp13 RNA helicase was shown having multiple domains and functions and ideal for targets for anti-viral drugs [30-32]. We confirmed the sequence similarities of L6 and L9 ribosomal proteins with Nsp13 proteins as well as Nsp2 has a relation to L1 etc supporting such view. We are much confused with the data but it appears that many non-structural proteins of Corona virus have significant homologies with the Ribosomal proteins and thus they could interfere with the ribosomal assembly correctly. Otherwords, Corona virus ribosomal proteins could interact with the Human ribosome preferably in the mitochondria to methylate as well as topological changes of mRNA or rRNA to alter the translational profile of host mRNA for protein synthesis or enhanced translation of viral structural genes [33]. Whatever the situation, we predicted that COX1 and COXII types proteins deficiency in mitochondria lead to low oxidative phosphorylation at the ETC and low ATP synthesis. This could cause many physiological abnormalities in host like sudden low blood pressure, coma and heart attack as found in many Corona patients. If ATP concentration in blood reached below 10 nM that could lead to blood clotting which was again shown recently in lung and brain of the many Corona patients in the Netherlands.
Mechanism of Corona virus pathogenesis is not known and no drug or vaccine yet approved today claiming every day>1000 peoples worldwide and within four months about 30 millions infections and >210000 deaths occurred mostly in the developed countries like USA, England, France, Italy and Spain. Asian and African countries also greatly have affected but due to early lock down technology death rate has restricted to hundreds only. Recently, Russia, Canada and Netherland also have affected to some extent. Sadly Corona pandemic created 100 millions jobless and worldwide panic and WHO has suggested that the pandemic may cause 20-50 millions death due to lack of food and sanitation. We must act to prevent the spread of Corona virus. Thus, the present bioinformatics analysis of Nsp9 and Nsp10 unknown proteins of Corona virus is important and may through some highlights to its control measures targeting those suspected ribosome antagonist and rRNAmethyl transferases [34,35].We previously showed that Nsp2 is a RNA topoisomerase, a great target for DNA topoisomerase inhibitors [36]. As well as Nsp13-16 may be other RlmE and Cfr-like methyl transferases although previous report has suggested Nsp13 as ATP-dependent RNA helicase [37-40].Many enzyme has multiple domains and thus they participate in multiple enzymatic activities as Nsp13 and Nsp9/10 which also have recombinase and transposase types domains [11]. Corona virus RNA genome replication is an ideal target and Nsp2 RNA topoisomerase has not been explored [41]. Alisporivir inhibited the SARS and MERS Corona viruses RNA replication and hydroxychloroquine was shown protective during Corona virus pathogenesis [42]. Interferon-αb therapy also has been started and under clinical trial [42-49].
We proposed a model where Nsp9, Nsp10 as well as previously reported Nsp13-16 have similarities to the ribosomal proteins and assembled in mitoribosome to methylate 21S and 12S rRNAs inhibiting protein synthesis. Low synthesis of COXI and COXII to types ETC enzymes cause low ATP synthesis which accelerates low blood pressure and heart attack. Further, very low ATP concentration also favours activation of platelets aggregation following factor XIII/IX-mediated thrombin-stimulated fibrin formation leading to blood clot in the lung and brain of COVID-19 patients. So targeting those RlmE, ErmDand RlmG types methyltransferases with phytochemicals, antisense, ribozyme and Crispr-Case6 technologies may be gene medicine for Corona virus pathogenesis. However, major focus for Corona-specific drug development was pinpointed to the ACE-2 receptor antagonist, RNA-dependent RNA polymerase (Nsp12), c3 protease (Nsp3) inhibitors (Hu et al.). Similarly, for vaccine development major focus molecules was studied likely spike protein of Corona virus. I have alternate thought like Nsp2 RNA topoisomerase and rRNAmethyltransferases. We have also designed peptide vaccine candidates and RT-PCR primers. We and other have developed many methods of toxic drug nanocarriersand plant extracts may be valuable source for superbugs and Corona virus control. Never the less, we clearly predicted the functions of Nsp9/10 proteins of Corona virus.
I thank Sir Narendra Modi, prime Minister of India for stimulation through his Mann Ki Baat programme to fight against COVID-19.
I have no conflict of interest to any company and any organization.
No patient data was used in this study. PDB Database and GenBank Database were analysed.
Citation: Chakraborty AK (2020) Corona Virus ORF1ab-Derived Nsp9 and Nsp10 Non-Structural Proteins have Homologies to S8/S10 Ribosomal Proteins as well as RlmG/ErmDrRNAMethyltransferases and may Inhibit Host Mitoribosome Assembly and Protein Synthesis. Virol Mycol. 9:186. DOI: 10.35248/2161-0517.20.09.186
Received: 30-Apr-2020 Accepted: 14-May-2020 Published: 21-May-2020 , DOI: 10.35248/2161-0517.20.09.186
Copyright: © 2020 Chakraborty AK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.