Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research - (2020)Volume 9, Issue 1
COVID-19 is a deadly infectious (+) stranded RNA Virus which produces a 7098AA length polyprotein that degraded into sixteen polypeptides in infected human cells. As the Coronavirus infections now pandemic claiming >300000 peoples worldwide, discovery of molecular target is great. The biological functions of the Coronavirus non-structural nsp2 protein is unknown. BLAST search is the best method to understand the function of unknown protein on the basis of sequence homology among the known protein amino acid sequence available in the Genbank Database. Few type I DNA topoisomerases show RNA Topoisomerase activity and such activity was found in Nsp2 protein by homology search with Vibrio haemolyticus Type I and type IV DNA topoisomerase as well as E. coli DNA gyrase and DNA primase. The RNA topoisomearse activity of COVID-19 needed to release the RNA-RNA knots and supercoils during (-) strand synthesis followed by sub-genomic mRNA synthesis. Such enzyme would be a target for DNA topoisomerases inhibitors used against bacterial infections and cancer. We concluded that Nsp2 protein is related to Type I and type IVDNA Topoisomerase of Vibrio haemolyticawhere NH2-terminas of topoVI has similarity to CO2H terminas of Nsp2 protein and CO2H domains of topoI has similarity stretches to the NH2-terminus of Nsp2 protein. Deep sea thermophillic bacteria like Desulfococcus sp and Marinobacter sp DNA topoisomerase I have 25%-30% homologies with stretches. Thus, Nsp2 protein is a RNA Topoisomerase of Coronavirus and is a strong candidate for drug design and vaccine development. Thirty amino acids length peptide of Nsp2 protein (H2N-LVN KFL ALC ADSIII GGA KLK ALNLGE TFV-CO2H) may be a good peptide vaccine against Coronavirus. We also designed nsp2F1 primer (5’-CCT GAT AGT CTT GCC GA-3’) and nsp2R1 primer (5’-GAG CAG TTT CAA GAG TGC GG-3’) for RT-PCR based diagnosis of COVID-19 infection. We are first to determine the function of Nsp2 protein of Coronavirus using bioinformatics approach.
RNA topoisomerase; Nsp2 protein; Covid-19; Novel target; Drug design; Peptide vaccine; RT-PCR
Coronaviruses (family Coronaviridae) are enveloped viruses with a largest positive sense, single-stranded RNA genome of 30kb [1]. On genetic and antigenic criteria, CoVs have been organised into three groups: α-CoVs, β-CoVs, and γ-CoVs [1,2]. Coronaviruses primarily infect birds, mammals and human, causing a variety of lethal respiratory diseases resembling the common cold, to lower respiratory tract infections such as bronchitis, pneumonia, and even severe acute respiratory syndrome (SARS). In recent years, coronaviral research must be augmented due to pandemic severe respiratory illnesses outbreaks claiming >500000 deaths [3]. Covid-19 virus enters cells through ACE2 receptor-mediated endocytosis (Figure 1). The receptor ACE2, was abundant in lungs AT2 alveolar epithelial cell as well as cells in the kidney, heart and blood vessels (Zhao, et al.). One of the known regulators of endocytosis is the AP2-associated protein kinase-1 implicated novel target for therapeutic intervention. Blockage of ACE-2 receptors prevents the aldosterone synthesis and thus deregulating Na+ absorption, blood pressure and normal renal function .The extremely large gene 1 covers 2/3 of the 30kb genome (separated into ORF1a and orf1b) encoding non-structural proteins involved in proteolytic processing of the gene 1 polyprotein products, virus genome replication, and sgmRNA synthesis (see, Table 1 for functions of corona virus Nsp proteins) (Figure 2) [4-6]. The rest 1/3 of the 3’end of the RNA genome encodes the structural spike glycoprotein (S), small envelope protein (E), membrane glycoprotein (M), and nucleocapsid protein (N) [5]. A number of other ORFs appearing to be virus group-specific, many apparently encoding non-structural proteins.These include ORF3a (7.7- kDa protein), ORF3b (27.7-kDa protein), and ORF4a (15.3-kDa protein) and ORF4b (10.2-kDa protein), ORF2a (32-kDa protein), ORF4 (17.8-kDa protein) and ORF5a (13.1-kDa protein) etc. [4], (accession nos.AY884001, DQ415912, KY674941, DQ415914, DQ415908, KT779555, KY674941, MH940245).DNA topoisomerases have roles in regulating replication, transcription and recombination [7-9]. DNA topoisomerases of viruses, bacteria and human have different structural assembly leading to drug development against variety of bacterial diseases targeting DNA topoisomerases. DNA gyrase is inhibited by fluoroquinolonesdrugs[10]. Eukaryotic DNA topoisomerases inhibitors like camptothecin, tenoposide and amsacrine are used successfully to treat variety of cancers [11,12]. So far no DNA topoisomerase was predicted in Coronavirus being a RNA virus. Neither function of Coronavirus polyprotein associated Nsp2 protein was discovered yet [13]. We are searching a DNA topoisomeraselike sequence homology in Coronavirus polyproteins hoping that conventional topoisomerase inhibitors may be useful to control its replication and transcription. Surprisingly, we detected a short homology of Nsp2 protein with Vaccinia DNA topoisomerase I. This lead to search for other DNA topoisomerases database and we found a 30% homology of Nsp2 protein with Vibrio haemolytica DNA topoisomerase IA. The validity of search however resides on the fact that few DNA topoisomerase I have been recently shown to have weak RNA swivelase and RNA knotting activities [8,14-17]. Type IA as well as Type III bacterial topoisomerases have shown to have RNA topoisomerase activities in vitro [15,18,19].
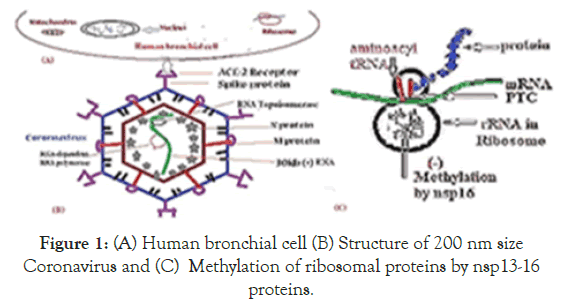
Figure 1. (A) Human bronchial cell (B) Structure of 200 nm size Coronavirus and (C) Methylation of ribosomal proteins by nsp13-16 proteins.
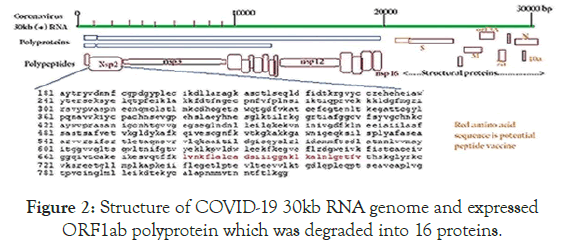
Figure 2. Structure of COVID-19 30kb RNA genome and expressed ORF1ab polyprotein which was degraded into 16 proteins.
| Name | AA length | Function |
|---|---|---|
| Nsp1 | 180 | Transcription factor Translational inhibitor [35,36] |
| Nsp2 | 638 | RNA topoisomerase Proinhibition1/2 binding & signalling Replication [37,38] |
| Nsp3 | 1945 | Papain-like Protease Polyprotein cleavage [39,40] |
| Nsp4 | 500 | DMV formatiom [3] |
| Nsp5 | 306 | 3C-lke Protease [41] |
| Nsp6 | 290 | Autophagosome and DMV formation [42] |
| Nsp7 | 83 | Cofators with Nsp8 and Nsp12 [43] |
| Nsp8 | 198 | DNA primase [44] |
| Nsp9 | 113 | RNA binding [45] |
| Nsp10 | 139 | Scaffold for Nsp14 and Nsp16 [6,46] |
| Nsp11 | 13 | Unknown |
| Nsp12 | 919 | RNA-dependent RNA polymerase [47,48] |
| Nsp13 | 601 | RNA helicase, 5’-triphosphatase [42,49] |
| Nsp14 | 527 | Exoribonuclease, capping MTase [50] |
| Nsp15 | 346 | Endoribonuclease [40,51,52] |
| Nsp16 | 298 | Capping MTase; RlmE MTase rRNA methylation [20,53] |
Table 1: Functions of corona polyprotein (7096aa) proteases cleaved polypeptides.
We have shown that Nsp16 RlmE methyltransferase methylates 21 S RRNA of human mitochondria causing the inhibition of mitochondrial protein synthesis and may stop oxidativep hosphorylation(20). Thus RNA topoisomerase also be a molecular target which changes the topology of + strand and _ strand RNA intermediates forming dimer structure unfavouable for mRNA synthesis.
The amino acid sequence of Nsp2 protein was given. Other structural proteins S, N, M, E were also seen in the (1/3 of the genome) were shown at the right. Nsp16 was aRlmEmethyltransferase [20].
In bacteria, there are two types of main DNA topoisomerases. Type I enzyme performs single-stranded nick and then after unwinding, liases the broken ends. Whereas Type II DNA topoisomerase of bacteria known as DNA Gyrase works by dsDNA cut and induces +vesuper coils on relaxed circular DNA and also decatenatesTrypanosomatidskDNA into free minicircles (Figure 3). In Achaea, another topoisomerase known as Reverse Gyrase which induces +ve superhelical turns on plasmid DNA. In eukaryotic organisms, there are multiple type I and type II DNA topoisomerases that control replication, transcription and recombination where as topoisomerase III, topoisomerase IVA/B and DNA gyrase of Vibrio sp. are well documented. Virus and bacteriophages also have such ubiquitious enzymes [21]. Both DNA topoisomerases can do DNA catenenes with monomeric plasmids in presence of histoneH1 or spermidine or DNA condensing proteins (Figure 3). DNA topoisomerase inhibitors are presented in Figure 4 being well known drug today. Thus, we predicted a DNA topoisomerase activity in Coronavirus similar to other viral and bacterial DNA topoisomerases. DNA topoisomerase III was located in many MDR conjugative plasmids and may involve in maintaining plasmid topology and conjugation. The Covid-19 vaccines are in clinical trials. The problem is in those cases antigenic load complicates due to many determinants and may induce allergic and/or reactogenic responses. Peptide vaccines use 20-30 amino acids of a antigenic region of a protein of a organism and very useful in protection for long time and now a attractive vaccine therapy protocol. Such peptide vaccine also requires particulate carriers for delivery and adjuventing. Covid-19 is a viral disease and nsp2 protein is unique and we devised three antigenic peptides to explore vaccination trials in animals [2].
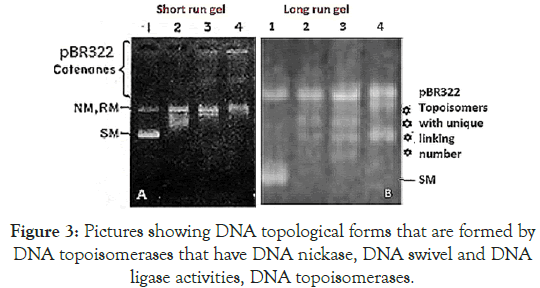
Figure 3. Pictures showing DNA topological forms that are formed by DNA topoisomerases that have DNA nickase, DNA swivel and DNA ligase activities, DNA topoisomerases.
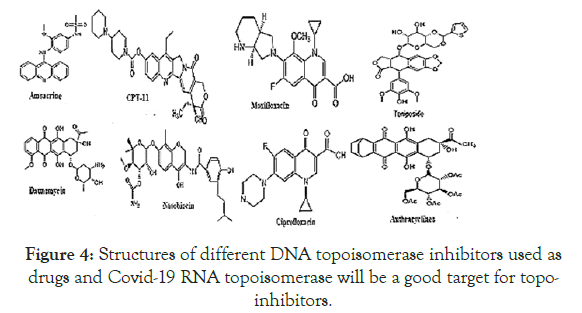
Figure 4. Structures of different DNA topoisomerase inhibitors used as drugs and Covid-19 RNA topoisomerase will be a good target for topoinhibitors.
Left panel: Lane 1, plasmid pBR322, lane 2+topoisomerase I, lane 3 and 4+topo II catenation in presence of spermidine and histone H1. Right panel: plasmid DNA, lane 2+topo I, lane 3, unique topoisomer of plasmid DNA+topo I with changes of linking number 1 and lane 4 + topo II with changes linking number of 2 [22,23].
We recommend testing topo-inhibitors against in vitro replication and transcription of Coronavirus [22].
The BLAST search was done using web portal www.ncbi.nlm.nih. gov/blast and retrive of covid-19 and other coronaviruses cDNA sequences were done using web portal www.ncbi.nlm.nih.gov/ nucleotide or protein. NCBI Primer Design Software was used for primer selection and Oligoanalyzer 3.2 software was used to analyze primer dimmer and hairpin structure. Multalin Software and CLUSTAL Omega Software were used to multiple align of protein sequences and NCBI BLAST seq-2 analysis portal used to analyze homology between two sequences. NCBI PubMed portal (www.ncbi.nlm.nih.gov/pubmed) used to retrieve references and papers. NEB DNA cutter software was used to restriction map teh DNA fragment.
Figure 5 showed the homology pattern of Vibrio haemolytica DNA topoisomerase I and Nsp2 protein of Coronavirus. Such results clearly demonstrated the origin of Nsp2 protein from DNA topoisomerases by recombination and mutations. However, the result lacks any good similarity and likely we may missed the real DNA topoisomerase I that may not yet deposited in the GenBank and PDB databases. So we continue our search on “DNA topoisomerase protein” in the GenBank database (www.ncbi.nlm. nih.gov/protein). In Figure 6 we demonstrated that among the many bacterial type I DNA topoisomerases, Marinobacter sp DNA topoisomerase has close homology to Nsp2 protein of Coronavirus. So, we searched many deep sea thermophillic bacteria DNA topoisomerase and found that Desulfurococcus sp and Pyrococcus sp deep sea thermophillic bacterial DNA topoisomerases have a relation to Nsp2 protein of Coronavirus (Figure 7). Interestingly, we found a relation with short homology of Nsp2 protein with DNA primase, gyrA and gyrB DNA gyrase subunits indicating the Nsp2 protein has acquired RNA/DNA binding, nickase and ligase domains from different regions of the related genes during its creation (Figure 8). Most astonishingly, when we compared the DNA topoisomerase IVA of Vibrio sp, we noticed the genesis of COOH-terminal half part of the Nsp2 occurred from the different parts of the Vibrio protein (Figure 9) where as Vibrio aligned to the NH2 terminus of the Nsp2 protein. So, we concluded that Nsp2 was derived from bacterial DNA topoisomerases and the changes occurred to obtain RNA specificity. RNA specificity is great being (+) stranded RNA virus and topological constraints may occur during (-) synthesis as dimmer RNA is not a mRNA to be transcribed for viral proteins in the host ribosome. There must be a dominant character of viral mRNA selection to push the cellular mRNAs as Coronavirus tropism is dominant claiming so far 300000 peoples worldwide. So, we confirmed that Nsp2 protein is a RNA topoisomerase with a distinct nature of DNA sequence and protein domains. So Nsp2 protein is a vital gene for RT-PCR mediated Coronavirus diagnosis and Nsp2 protein also is an ideal candidate for vaccine development. Keeping that mind we also designed a pair of primers for RT-PCR detection and such primers are specific for COVID-19 (Figure 10A). The restriction pattern of the PCR fragment has also determined (Figure 10B) and the 1% agarose gel electrophoresis pattern also has presented for common worker using three well known restriction endonucleases (Figure 10B). We also designed three hydrophobic amino acid rich peptides (nsp2V1, nsp2V2 and nsp2V3) for vaccination in human. We thought that nsp2V3 could be a good peptide vaccine candidate with two phenyl alanine, five leucine, two valine, three isoleucine and two asparagine (n) for N-glycolysation and one serine (s) for O-glycolysation (Figure 11). That way we confirmed the utility of Coronavirus Nsp2 protein being a RNA topoisomerase and target for drug and vaccine development and Nsp2 enzyme is indispensible enzyme of Coronavirus life cycle (Figure 12). From the homology patterns of DNA topoisomerase I, DNA topoisomerase IVA Gyrase A subunit, Gyrase B subunit, DNA primase and RNA ligase we have determined the different domains with ATPase activity, Nicase activity, ligase activity and DNA/RNA binding activities (Figure 13). We have also searched the Nsp2 proteins of other RNA viruses ad negligible sequence similarities were observed. Rotavirus seven segmented ds-RNA genome (total 17kb) associated Nsp2 nonstructural RNA binding protein (protein id. ADF57890) has some similarities at the C-terminal RNA Swivelase domain (494aa-579aa) of Nsp2 RNA topoisomerase of Coronavirus which has gyrA and DNA primase homology stretches. Commensurate with rotavirus Nsp2 has ATPase activity, 98aa-135aa region of minor homology detected in Coronavirus Nsp2 protein (24%). Similarity 889aa long Nsp2 protein of sense single stranded Sindbis fever virus (11.7kb RNA genome) has different 20-25% similarity stretches at the RNA nicking region. Nsp2 protein of Hepatitis C virus ( 9.6kb, sense RNA) belong to polyprotein (3033aa; BAJ07247) is small (194aa) but has some similarity to Nsp2 of Coronavirus at the gyrA homology region (400aa-457aa). That way we have tried to compare the homology of different RNA viruses Nsp2 proteins which have reported with RNA binding or RNA helicase or endoribonuclease activities but none has over 25% similarities stretches.

Figure 5. Sequence similarity between Vibriohaemolytica DNA topoisomerase IA and Corona Virus polyprotein associated nsp2 protein using Multalin software.
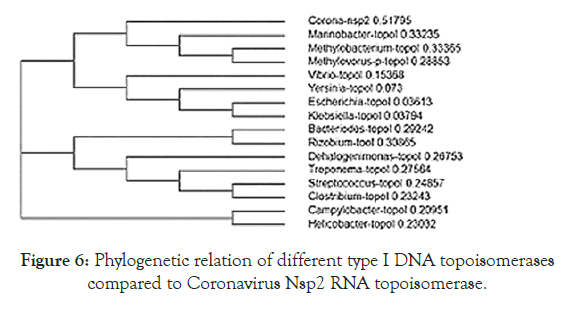
Figure 6. Phylogenetic relation of different type I DNA topoisomerases compared to Coronavirus Nsp2 RNA topoisomerase.
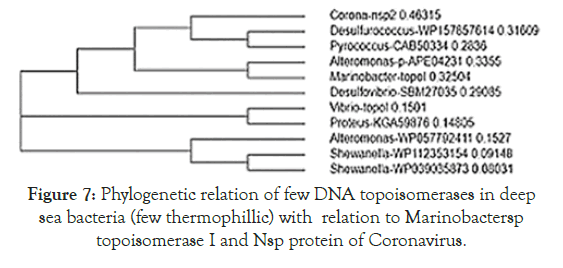
Figure 7. Phylogenetic relation of few DNA topoisomerases in deep sea bacteria (few thermophillic) with relation to Marinobactersp topoisomerase I and Nsp protein of Coronavirus.

Figure 8. Similarity between Nsp2 protein and E. coli DNA primase (A); DNA GyrA subunit (B); GyrB subunit (C) and T4 RNA ligase (D).
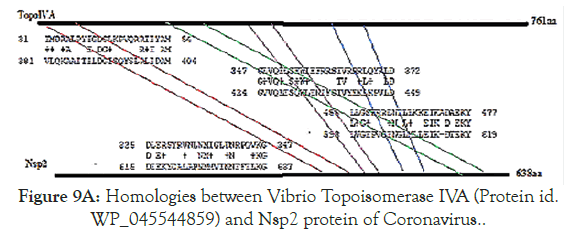
Figure 9A. Homologies between Vibrio Topoisomerase IVA (Protein id. WP_045544859) and Nsp2 protein of Coronavirus.

Figure 9B. Homologies between Vibriosp topoisomerase IA and Nsp2 protein of Coronavirus.

Figure 9C. Seq-2 BLAST amino acid sequence homology between chimera VibriotopoIA (protein id.WP_109168522; 481-876aa) + topoIVA (protein id.WP_045544859; 1-480aa) and Nsp2 protein of Coronavirus.
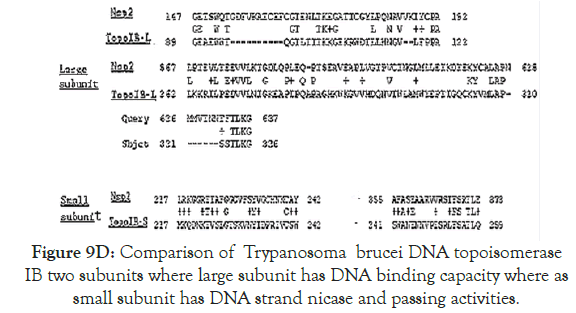
Figure 9D. Comparison of Trypanosoma brucei DNA topoisomerase IB two subunits where large subunit has DNA binding capacity where as small subunit has DNA strand nicase and passing activities.
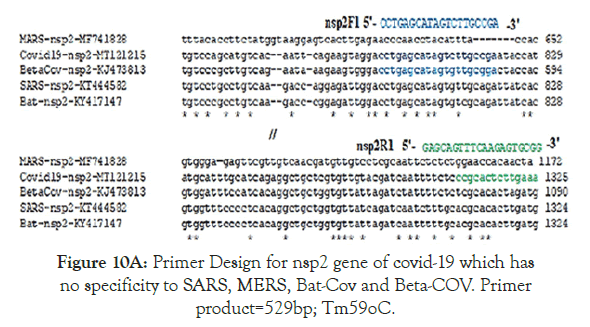
Figure 10A. Primer Design for nsp2 gene of covid-19 which has no specificity to SARS, MERS, Bat-Cov and Beta-COV. Primer product=529bp; Tm59oC.
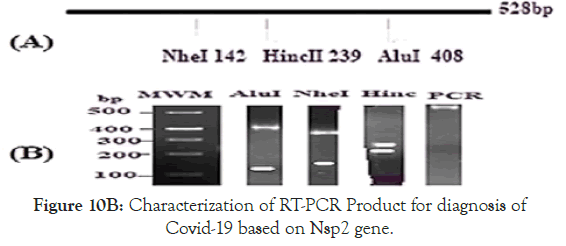
Figure 10B. Characterization of RT-PCR Product for diagnosis of Covid-19 based on Nsp2 gene.
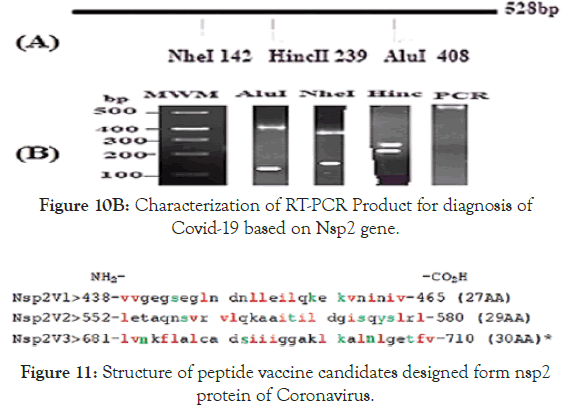
Figure 11. Structure of peptide vaccine candidates designed form nsp2 protein of Coronavirus.
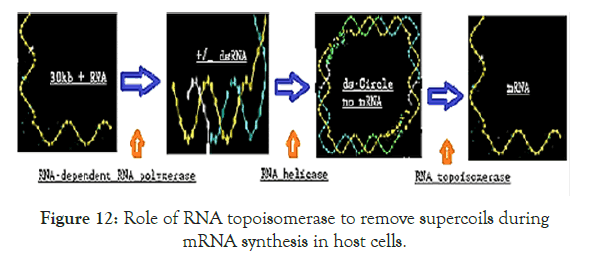
Figure 12. Role of RNA topoisomerase to remove supercoils during mRNA synthesis in host cells.
It appears Desulfococcus sp DNA topoisomerase I has close relation to Nsp2 protein of Coronavirus than Marinobactersp as discussed above.
The stretches of similarity supports the view that RNA binding domains must be there in RNA primase and if Nsp2 is a RNA topoisomerase, then some stretches of homology with Nsp2 must as was shown here. DNA primase sequence (protein id. AVX50839) was obtained from E. coli MDR plasmid pEsCo-36073cz (accession no. MG252895). The specificity is there as DNA topoisomerase III of E. coli plasmid pMG252A (accession no. MK733575; protein id. QCO89636) has no such homology. Homology region of DNA GyrA of Vibrio sp (protein id. WP_021705456) and Nsp2 protein of Corona virus. Homology region of GyrB with Nsp2 protein of Coronavirus. The region of Nsp2 aligned with GyrA (444-562 AA) is different with GyrB (89-136AA) reflecting the domain functions could be mapped knowing the domains of GyrA and GyrB subunits. Region 461-527 has homology with DNA primase coinside with GyrA homology but not the 2nd homology region at 287-315AA of nsp2 protein. Seq-2 BLAST homology between T4 RNA ligase A chain and Nsp2 protein of Coronavirus. Idea is as the substrate is RNA in RNA ligase and thus will be a homology region in RNA topoisomerase Nsp2 protein. We tried to find a >50% homology but failed. That indicated Nsp2 protein of Coronavirus was originated by multiple recombinational and mutational events.
It appears four regions of the Vibrio topoIVA protein were assembles into the COOH terminas of Nsp2 protein suggesting many recombination and mutations events were happened to make Nsp2 protein (Figure 9A).
It is proved that recombination between Vibrio sp topoisomerase I and topoisomerase IV may be primary step for the generation of Coronavirus RNA topoisomerase and further mutations and rearrangement have occurred to get specificity to COVID-19. However, Desulfurococcus sp or Marinobacter sp or any other missed deep vent sea bacteria or bacteriophages DNA topoisomerases (as discussed in Figures 8 and 9) may be the originator of Coronavirus RNA topoisomerase (Figure 9B).
This helped to understand domains of nsp2 RNA topoisomerase where amino acids 567-637 has RNA binding, amino acids 217- 257 has catalytic nicase/swivalse and aminoacids 147-192 has ATPase activities. Figure 10B demonstrated the map of the RTPCR fragment as well as restriction pattern of 1% agarode gel ran two hrs at 60V in presence of 1XTAE and o.5 o.5μg/ml ethidium bromide [20].
Vaccination is the best remedy for infectious ver small virus particles like Corona virus and we designed few peptide vaccines (Figure 11). Red is hydrophobic amino acids (l=Leucine, v=Valine, i=Isoleucine and f=Phenylalanine) and green is amino acids that could be glycosylated, phosphorylated or acetylated (s=Serine, t=Threonine, k=Lysine and y=Tyrosine). Numbering is based on polyprotein ORF1ab (protein id.QII57165) of Covid-19. We anticipated to make an recombinant vector under Pol-III promoter to express peptide vaccine and injection of such plasmid into muscle under actin promoter will boost permant protection against coronavirus [23-25]. In truth, 100 of such peptide vaccines are under clinical trials.
We also have predicted the functions of a RNA Topoisomerase activity of Nsp2 protein. Without RNA topoisomerase no sufficient Cononovirus mRNA for synthesis of structural proteins and thus no virus propagation causing pathogenesis. So nano-carrier mediated delivery of antisense or ribozyme to nsp2 will be lethal to Coronavirus. Interestingly, we mapped the homology domains of known topoisomerases with Nsp2 RNA topoisomerase as described in Figure-13 on the basis of data analysis described in Figures 8-11.
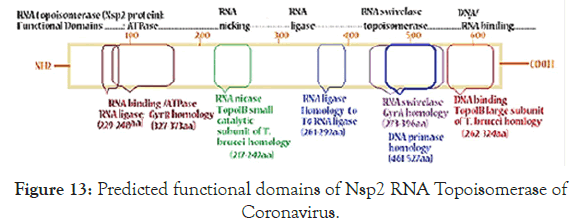
Figure 13. Predicted functional domains of Nsp2 RNA Topoisomerase of Coronavirus.
We also have predicted the functions of a RNA Topoisomerase activity of Nsp2 protein.Without RNA topoisomerase no sufficient Cononovirus mRNA for synthesis of structural proteins and thus no virus propagation causing pathogenesis. So nano-carrier mediated delivery of antisense or ribozyme to nsp2 will be lethal to Coronavirus. Interestingly, we mapped the homology domains of known topoisomerases with Nsp2 RNA topoisomerase as described in Figure-13 on the basis of data analysis described in Figures 8-11.
No function of Nsp2 protein of COVID-19 has confirmed yet. We first described Nsp2 protein as RNA topoisomerase being very similar to DNA topoisomerase IA structure and function. DNA topoisomerases have ubiquitous role in cell physiology and marker of variety of genetic diseases. We predicted that proteome homeostasis of bronchial cells was deregulated during Coronavirus infection and RNA topoisomerase must have role in preference of viral protein synthesis [26].We searched many Nsp2 non-structural proteins implicated as RNA binding proteins and we assigned such enzymes as RNA topoisomerase. Sindbis virus, HCV and Rotavirus Nsp2 have little similaritiesbut more analysis to be done. Complex nature of RNA and protein structures in ribozyme mediated gene regulation and the circRNAs in Ribozyme has profound roles in modulating transcriptosome signatures [19,27]. Vital protein synthesis regulators in eukaryotes would be modulated by animal viruses and capping 2’-O- Ribose methyltransferase protects viral mRNA [28]. Many non-coding RNAs regulate gene expression [29]. As for example, expression of Nicase and caspase9 caused the inhibition of HBV replication and HaSV replication [30-32]. DNA topoisomerase I is well target for Trypanosomatid drugs [23,33]. Expression of antisense to Nsp2 RNA topoisomerase may inhibit Coronavirus replication where as small ribozyme could be cloned in retroviral or retrotransposon mediated gene delivery systems targeting Nsp2 protein of Coronavirus [24,25]. Bioinformatics approach is a good method to predict cellular process of COVID-19 molecular biology; otherwise COVID-19 work needs high Biosafety standard laboratory [29]. We vision to identify COVID-19 RNA topoisomerase specific drugs whereas it was very good to find action of conventional topoisomerase inhibitors as molecular target (Figure 3). We also designed three peptide vaccines and good to see its effects on animals and then in human [2].We satisfy the valuable urgent need for controlling Coronavirus spread claiming now >120000 in the developed countries like USA and UK. The present report will stimulate research in the developed nations having Biosafety-3 level laboratories to work with live Coronavirus. We also actively working with TLC-purified heterogeneous phytoantibiotics isolated from bark and root of Indian medicinal plants like Suregada multiflora, Cassia fistula, Shorea robusta and Jatropha gossypiifolia to control infectious diseases [34]. Our hope to test such active chemicals on Coronavirus replication and we are in communication to the COVID-19 experts worldwide.
Nsp2 protein of Coronavirus among the type-1A and type IVA DNA topoisomerases of Vibrio sp and Desulfurococcas sp as well as E. coli DNA primase and T4 RNA ligase to conclude that Nsp2 was a RNA topoisomerase that might regulate the RNA genome topology. The work is needed very sophisticated laboratory to prove our hypothesis but our approach is considerably sound and we are very sure that Nsp2 has RNA topoisomerase activities related to a Type I DNA topoisomerases. We designed COVID-19 based RT-PCR primers and also peptide vaccines for the diagnosis and vaccine therapy for human Coronavirus. COVID-19 infections have very much community spread in the China, USA, Italy, Spain, France and UK but Canada, Russia, Iran, India and other South Asian Countries also have faced COVID-19 infections which largely has controlled by Lock Down Technologies. “Test Test and Test” was proclaimed by WHO Director General and Lock Down who become positive. COVID-19 Virologist and Head of Infectious Diseases at National Institute of Health (NIH, USA) Dr Anthony S Fauci alarms, COVID-19 is more deadly than SARS and MERS Corona viruses. So, vaccination protocol must be developed quickly.
I thank Dr Bidyut Bandhopadhyay, Principal of OIST for his interest in Coronavirus research.
I have no conflict of interest
No patient data was used in the study. It is lock down research at home using a single computer with few DNA and protein analysis softwares and NCBI databases.
Citation: Chakraborty AK (2020) Coronavirus Nsp2 Protein Homologies to the Bacterial DNA Topoisomerase I and IV Suggest Nsp2 Protein is an Unique RNA Topoisomerase with Novel Target for Drug and Vaccine Development.Virol Mycol. 9:185. DOI: 10.35248/2161-0517.20.09.185
Received: 30-Apr-2020 Accepted: 09-May-2020 Published: 15-May-2020 , DOI: 10.35248/2161-0517.20.09.185
Copyright: © 2020 Chakraborty AK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.