Chemotherapy: Open Access
Open Access
ISSN: 2167-7700
ISSN: 2167-7700
Research Article - (2024)Volume 12, Issue 4
Objective: This study aims to comprehensively explore the comorbidity mechanisms of eczema and Atopic Dermatitis (AD) through the integrated application of Weighted Gene Co-Expression Network Analysis (WGCNA), machine learning, and bioinformatics methods. The goal is to identify potential therapeutic targets and provide a profound understanding of these two skin conditions.
Methods: We conducted Weighted Gene Co-Expression Network Analysis (WGCNA) and Differential Gene Expression (DEGs) analysis using the Gene Expression Omnibus (GEO) databases GSE6012, GSE14550, GSE32924, and GSE120721. By intersecting the results, we identified 22 genes that are commonly expressed in eczema and atopic dermatitis. Functional enrichment analyses, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), disease prediction, and Protein-Protein Interaction (PPI), were performed on these genes. Subsequently, key hub genes— CCL18, GZMB, and IRF7—were selected using random forest in machine learning and the cytohubba function in Cytoscape. We further explored the relationships between these three genes and immune cells through CIBERSORT and Single-Sample Gene Set Enrichment Analysis (ssGSEA), establishing a mRNA-miRNA-lncRNA ceRNA regulatory network.
Results: Enrichment analysis revealed that these 22 genes are predominantly involved in processes such as cytokine- mediated signaling pathways, leukocyte migration, leukocyte chemotaxis, neutrophil chemotaxis, and humoral immune responses. Additionally, they are associated with various skin-related diseases, including atopic dermatitis. Machine learning and network analysis confirmed CCL18, GZMB, and IRF7 as key genes. Immunoinfiltration analysis further elucidated the associations of these genes with different immune cells.
Conclusion: The results of this study suggest that CCL18, GZMB, and IRF7 may serve as potential biomarkers for eczema and atopic dermatitis, holding significant clinical prospects. These genes play vital roles in various biological processes and immune regulation, providing robust support for unraveling the comorbidity mechanisms of these two skin conditions. This discovery offers essential clues for future exploration of therapeutic targets and clinical interventions, potentially advancing research in the treatment of eczema and atopic dermatitis.
Eczema; Atopic dermatitis; Bioinformatics; Weighted Gene Co-Expression Network Analysis; Machine learning
Eczema is a common skin disorder and is classified as an allergic disease. The occurrence of eczema is influenced by various internal and external factors, with its primary clinical features including intense pruritus and dermatological manifestations [1]. For patients with severe inflammation, symptoms may spread throughout the body.
Due to the relatively slow progression of eczema, exposure to sensitizing factors during treatment may lead to recurrent outbreaks [2]. Atopic Dermatitis (AD) is a chronic inflammatory skin disease affecting millions worldwide, characterized by intense itching and eczematous lesions in typical anatomical locations [3-4]. Both eczema and atopic dermatitis fall within the classification of chronic dermatological conditions [5]. In clinical settings, these terms are occasionally employed interchangeably owing to their shared symptoms; however, it is important to note that Atopic Dermatitis is technically a subset of eczema. Atopic Dermatitis is associated with International Classification of Diseases, Ninth Edition (ICD-9) code 691.8 and International Classification of Diseases, Tenth Edition (ICD-10) code L20.x. However, the term eczema is associated with ICD-9 code 692.9 and ICD-10 code L30.9 [6]. The pathogenesis of these skin diseases is highly complex, lacking specific therapeutic approaches5, involving aspects such as immune regulation, inflammatory responses, epidermal dysfunction, and skin microbiome abnormalities [7]. Aberrant regulation of gene expression at the biological level has emerged as a pivotal determinant in the pathogenesis of eczema and Atopic Dermatitis. In recent times, the extensive utilization of bioinformatics techniques has facilitated a more exhaustive and organized comprehension of the molecular mechanisms underpinning these afflictions.
By employing a combination of Weighted Gene Co-Expression Network Analysis (WGCNA) and machine learning methodologies, our objective is to elucidate the underlying co-morbid mechanisms associated with eczema and atopic dermatitis, while exploring potential therapeutic targets. This research trajectory not only enhances our comprehension of the pathogenesis of these diseases, but also furnishes substantial bioinformatics backing for forthcoming treatment approaches. By employing a rigorous bioinformatics analysis, our study aims to elucidate the genetic associations between eczema and Atopic Dermatitis, thereby enhancing our comprehension of these prevalent dermatological disorders and facilitating further investigations into treatment modalities. The specific workflow diagram is shown in Figure 1.
Figure 1: The pathways of bacterial translocation through the Gastrointestinal Tract (GIT) epithelium. Note: In healthy individuals, antimicrobial molecules as part of the innate and immune adaptative response are secreted from intestinal epithelial cells and paneth cells and kill enteric bacteria. Drug suppresses the expression of these molecules resulting in intestinal bacterial overgrowth and dysbiosis. This might contribute to bacterial translocation observed after drug that also exert a direct effect on the intestinal microflora. There are five pathways for bacterial translocation: 1) Endocytosis by enterocytes; 2) Absorption (transcytosis) by M cells; 3) Passage by persorption through gaps left in the epithelium after the loss of enterocytes (apoptosis); 4) Paracelullar translocation; 5) Transcellular translocation.
Chip data collection and workflow
Utilizing the GEO database for the extraction of gene expression profiles. GSE6012 comprises gene expression profiles from 20 samples, including 10 samples from patients with active atopic dermatitis and 10 healthy control skin samples. GSE57225 includes 62 samples, consisting of 17 healthy control skin samples, 23 samples from eczema-affected skin, and 22 samples from psoriasis- affected skin. GSE32924 contains 33 samples, encompassing 8 healthy control skin samples and 25 samples from atopic dermatitis- affected skin.
GSE120721 comprises 52 samples, including 22 healthy control skin samples and 30 samples from atopic dermatitis-affected skin. GSE175438 consists of 31 samples, with 10 healthy control skin samples, 12 samples from atopic dermatitis-affected skin, and 12 samples from psoriasis-affected skin. Lastly, GSE168694 includes 8 samples, comprising 4 normal Peripheral Blood Mononuclear Cell (PBMC) samples and 4 atopic dermatitis PBMC samples.
Bioinformatics analysis
Differential gene expression analysis: Download normalized expression matrices of microarray data from four datasets and represent them using boxplots (created with the boxplot function). Then, annotate the probes using the annotation files from the respective datasets. Validate the reproducibility of the data through Principal Component Analysis (PCA) and create a PCA plot using the 'plot' function in R. Utilize the 'limma' package in R to identify differentially expressed genes (DEGs) by considering genes with adjusted p-values less than 0.05 and absolute Fold Change (FC) greater than 1. Generate heatmaps, volcano plots, and boxplots using the 'heatmap,' 'ggplot2,' and 'plot' functions in R software (version 4.2.1).
Weighted gene co-expression network analysis
WGCNA is a vital tool in bioinformatics analysis and has been widely employed for the analysis of associations between traits and genes [8]. We employed the R software package "WGCNA" to construct co-expression networks using gene expression data from GSE6012, GSE57225, GSE32924, and GSE120721 as input, with traits being either eczema/normal or atopic dermatitis/normal. Initially, sample clustering was performed using the hclust function, removing outlier samples, with distance calculated using the "method=average" parameter. Subsequently, an optimal soft threshold was determined to obtain a scale-free network. The dynamic tree- cutting algorithm was then applied to segment modules, and modules associated with eczema or atopic dermatitis were identified using Pearson correlation analysis. To comprehensively consider genes related to eczema and atopic dermatitis as identified by WGCNA and DEGs, a Venn diagram was employed to analyze common genes across the four datasets. In the end, a set of 22 genes associated with both conditions was identified.
Protein-Protein Interaction Networks (PPI)
The Protein-Protein Interaction (PPI) network among the 22 genes was visualized using the STRING database. Utilizing the cytohubba plugin in Cytoscape software, we obtained the PPI network for the top 10 hub genes, highlighting key genes within the interaction network.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
Conducting Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using R software, where corrected p-values less than 0.05 indicate statistically significant differences. The prediction of disease association for the 22 genes is being performed through the Metascape website.
Machine learning for selecting key genes
In the realm of machine learning, the Random Forest (RF) model was employed, utilizing the randomForest function within the random forest R package [9]. Gene selection was conducted on four datasets, namely GSE6012, GSE57225, GSE32924, and GSE120721. Subsequently, an intersection analysis was performed with the top 10 hub genes obtained from the cytohubba plugin in Cytoscape software. Ultimately, three key genes CCL18, GZMB, and IRF7 were identified.
Immune infiltration analysis
The Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm was employed to calculate the proportions of immune cell types, including T cell subtypes, B cells, and macrophages, in each sample [10].
Subsequently, the ssGSEA algorithm from the R package "GSVA" was utilized to assess the immune activity of each sample. Following the completion of CIBERSORT and ssGSEA calculations, correlation analyses among immune cell types were conducted, and differences in immune infiltration between two groups were analyzed. These analyses aimed to investigate the associations between immune infiltration and the three key genes.
Construction of an lncRNA-Associated ceRNA Regulatory Network
Utilizing TargetScan 8.0, miRWalk, and Tarbase v8 databases, miRNA prediction was performed for CCL18, GZMB, and IRF7. To enhance accuracy, an intersection analysis was carried out with the upregulated miRNAs from the GSE175438 dataset, resulting in a more consistent miRNA set. Subsequently, the obtained miRNA set underwent enrichment analysis using the miEAA database, ultimately identifying 11 miRNAs participating in all pathways. Employing the miRNet 2.0 database, lncRNA prediction was conducted for these 11 miRNAs. To enhance accuracy, the predicted lncRNAs were intersected with the upregulated lncRNAs from the GSE168694 dataset, culminating in the construction of the final ceRNA regulatory network involving lncRNA-miRNA- mRNA.
Statistical analysis
All statistical analyses were conducted using R software (version 4.1.3). The Wilcoxon test was employed to compare differences in continuous variables, and the Pearson correlation coefficient was utilized to assess the correlation of continuous variables. Data visualization was performed using the "ggplot2" package in R. All tests were two-sided, with a significance threshold set at p<0.05. The levels of significance are denoted as follows: *p<0.05, **p<0.01, ***p<0.001.
Selection of genes associated with eczema and atopic dermatitis
The expression matrices of the GSE6012, GSE57225, GSE32924 and GSE120721 datasets were normalized, and the distribution trends in the boxplots were generally linear. To assess the intra- group data repeatability, Principal Component Analysis (PCA) analysis was performed on the four datasets. The results indicated good repeatability for all datasets except GSE32924 (Figures 2A- 2H).

Figure 2: A-D) The normalized expression matrices; E-H) Principal component analysis (PCA) plots of the GSE6012, GSE32924, GSE57225 and GSE120721 datasets.
After filtering based on the adjusted threshold |log2(FC)|>1 and p-value<0.05, 566 differentially expressed genes Diethylene glycol (DEGs) were identified in the GSE6012 dataset (323 upregulated, 243 downregulated), 907 DEGs in the GSE57225 dataset (539 upregulated, 368 downregulated), 901 DEGs in the GSE32924 dataset (498 upregulated, 403 downregulated), and 1094 DEGs in the GSE120721 dataset (562 upregulated, 532 downregulated). The volcano distribution plots of DEGs in these four datasets are shown in (Figures 3A-3L). Additionally, WGCNA was conducted on these four datasets, and display the hierarchical clustering dendrograms of genes and the heatmap of gene modules correlated with clinical features, respectively. The dynamic tree- cutting algorithm was employed to identify gene modules in four different datasets, namely GSE6012, GSE57225, GSE32924, and GSE120721, resulting in the identification of 91, 29, 24, and 12 gene modules. Modules exhibiting a correlation coefficient (R) greater than 0 and a p-value less than 0.05 in were deemed to be associated with eczema and atopic dermatitis. Consequently, the genes encompassed within these modules were considered to be implicated in the pathogenesis of eczema and atopic dermatitis.
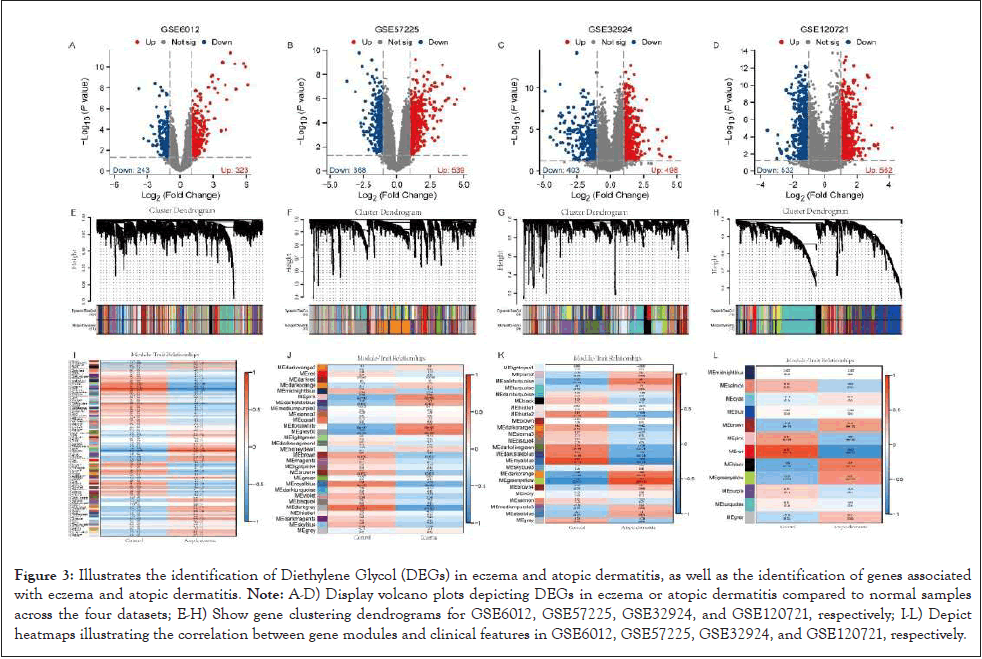
Figure 3: Illustrates the identification of Diethylene Glycol (DEGs) in eczema and atopic dermatitis, as well as the identification of genes associated with eczema and atopic dermatitis. Note: A-D) Display volcano plots depicting DEGs in eczema or atopic dermatitis compared to normal samples across the four datasets; E-H) Show gene clustering dendrograms for GSE6012, GSE57225, GSE32924, and GSE120721, respectively; I-L) Depict heatmaps illustrating the correlation between gene modules and clinical features in GSE6012, GSE57225, GSE32924, and GSE120721, respectively.
Enrichment analysis and disease prediction of core genes associated with eczema and atopic dermatitis
Following the analysis of Differentially Expressed Genes (DEGs) and Weighted Gene Co-Expression Network Analysis (WGCNA) on the GSE6012, GSE57225, GSE32924, and GSE120721 datasets, we obtained a gene set associated with eczema and atopic dermatitis. By intersecting the upregulated genes from DEGs in the four datasets and genes related to eczema and atopic dermatitis from WGCNA analysis, we finally identified a set of 22 genes. The chromosomal distribution of these 22 genes is depicted. The expression heatmaps for these 22 genes in GSE6012, GSE57225, GSE32924, and GSE120721 are presented in (Figure 4A-4L). To explore potential interactions among these genes, we constructed a protein-protein interaction (PPI) network for the 22 genes in the string database, and the results of the PPI network are illustrated.
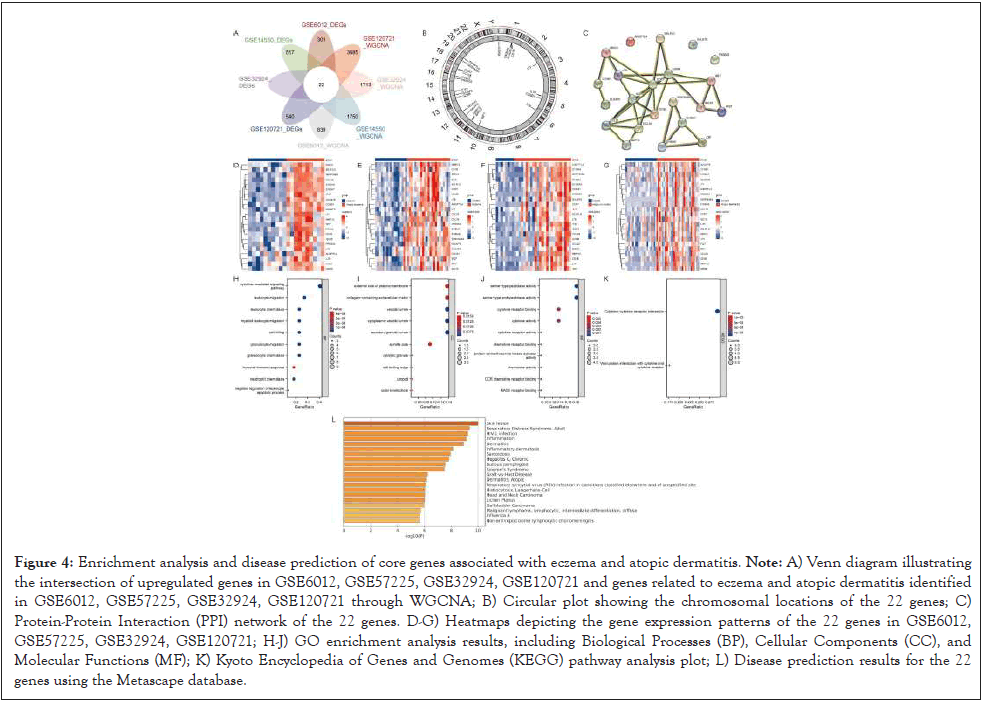
Figure 4: Enrichment analysis and disease prediction of core genes associated with eczema and atopic dermatitis. Note: A) Venn diagram illustrating the intersection of upregulated genes in GSE6012, GSE57225, GSE32924, GSE120721 and genes related to eczema and atopic dermatitis identified in GSE6012, GSE57225, GSE32924, GSE120721 through WGCNA; B) Circular plot showing the chromosomal locations of the 22 genes; C) Protein-Protein Interaction (PPI) network of the 22 genes. D-G) Heatmaps depicting the gene expression patterns of the 22 genes in GSE6012, GSE57225, GSE32924, GSE120721; H-J) GO enrichment analysis results, including Biological Processes (BP), Cellular Components (CC), and Molecular Functions (MF); K) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis plot; L) Disease prediction results for the 22 genes using the Metascape database.
Enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), were performed on these 22 genes. The results indicated that the most enriched pathways in GO activities included cytokine- mediated signaling pathways, leukocyte migration, leukocyte chemotaxis, bone marrow leukocyte migration, cell killing, granulocyte migration, granulocyte chemotaxis, humoral immune response, negative regulation of neutrophil apoptosis, and regulation of leukocyte apoptotic process. In the KEGG analysis, these 22 genes were involved in cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor pathway. Disease prediction using the Metascape database revealed associations with various conditions such as skin injuries, inflammation, dermatitis, inflammatory skin diseases, pemphigus vulgaris, atopic dermatitis, and lichen planus.
Machine learning and network analysis unveil key genes in eczema and atopic dermatitis
Through the integration of DEGs and WGCNA, a set of 22 genes was identified. Subsequently, machine learning techniques were employed to perform random forest analysis on the expression profiles of these genes in four datasets (GSE6012, GSE57225, GSE32924, and GSE120721) (Figures 5A-5N). Additionally, the cytohubba plugin in Cytoscape software was utilized to determine the top 10 hub gene. By intersecting the genes selected by random forest with the top 10 hub genes, three key genes, namely CCL18, GZMB, and IRF7, were ultimately identified.
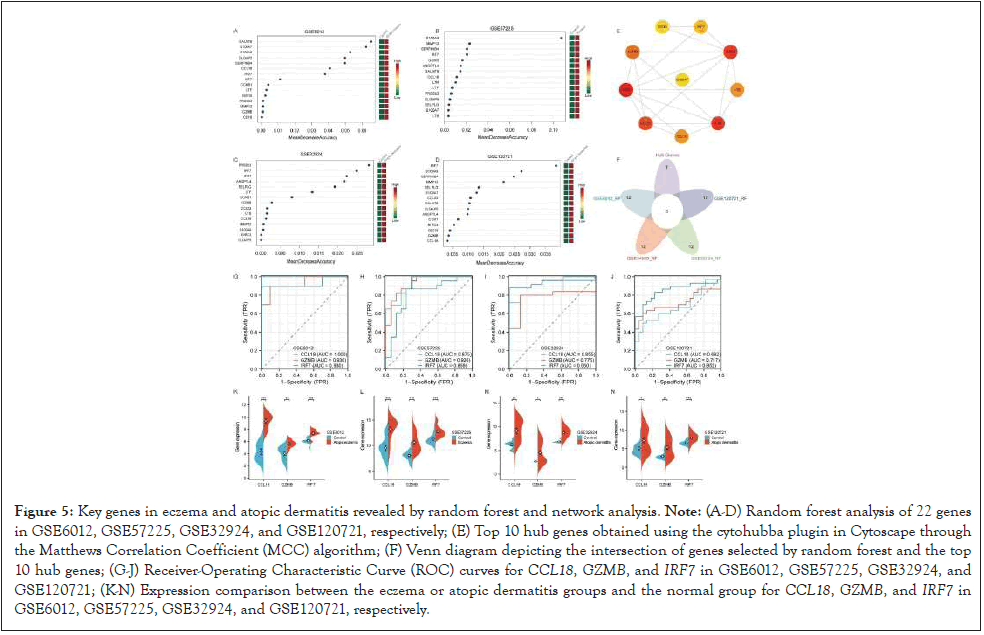
Figure 5: Key genes in eczema and atopic dermatitis revealed by random forest and network analysis. Note: (A-D) Random forest analysis of 22 genes in GSE6012, GSE57225, GSE32924, and GSE120721, respectively; (E) Top 10 hub genes obtained using the cytohubba plugin in Cytoscape through the Matthews Correlation Coefficient (MCC) algorithm; (F) Venn diagram depicting the intersection of genes selected by random forest and the top 10 hub genes; (G-J) Receiver-Operating Characteristic Curve (ROC) curves for CCL18, GZMB, and IRF7 in GSE6012, GSE57225, GSE32924, and GSE120721; (K-N) Expression comparison between the eczema or atopic dermatitis groups and the normal group for CCL18, GZMB, and IRF7 in GSE6012, GSE57225, GSE32924, and GSE120721, respectively.
Illustrate the gene expression group comparison plots for these three genes in GSE6012, GSE57225, GSE32924, and GSE120721. Clearly, the expression of these three genes is significantly higher in the eczema or atopic dermatitis groups compared to the normal group (p<0.05). The ROC curves created using the data from the eczema or atopic dermatitis groups and the normal group. The results indicate that CCL18 has an AUC greater than 0.85 in all datasets except GSE120721. GZMB has an AUC greater than 0.92 in GSE6012 and GSE57225, and an AUC greater than 0.7 in GSE32924 and GSE120721. Additionally, IRF7 has an AUC greater than 0.85 in all four datasets. In summary, these three genes demonstrate significant diagnostic and predictive value in eczema and atopic dermatitis.
Immune infiltration analysis
By comparing the differences between eczema or atopic dermatitis tissues and control groups, we employed the CIBERSORT algorithm and Gene Set Enrichment Analysis (ssGSEA) algorithm to delve into the comprehensive landscape of immune infiltration in eczema or atopic dermatitis. In samples from GSE57225 and GSE120721, we illustrated the proportions of 22 immune cells, as shown in (Figures 6A-6H). Histogram distributions of immune cell contents are visible. In GSE57225, the content of T cells CD4+ naive, activated memory CD4+ T cells, and M1 macrophages in the eczema group significantly increased compared to the normal group, while the content of plasma cells and resting mast cells decreased. In GSE120721, the atopic dermatitis group exhibited a higher proportion of activated memory CD4+ T cells, while the proportions of regulatory T cells, activated NK cells, and resting mast cells were lower. Demonstrate the correlations between immune cells in the GSE57225 and GSE120721 datasets. In GSE57225, activated memory CD4+ T cells showed a positive correlation with initial CD4+ T cells, CD8+ T cells, and resting dendritic cells, and a negative correlation with resting mast cells. In the GSE120721 dataset, activated memory CD4+ T cells were positively correlated with M2 macrophages and γδT cells, and negatively correlated with neutrophils, resting mast cells, M0 macrophages, activated NK cells, regulatory T cells, plasma cells, and memory B cells. Additionally, Pearson correlation coefficients revealed relationships between immune cell abundance and hub gene expression. In eczema, the three genes showed a positive correlation with activated memory CD4+ T cells and a negative correlation with resting mast cells. Furthermore, GZMB and IRF7 showed a positive correlation with M1 macrophages, while CCL18 and GZMB showed a positive correlation with initial CD4+ T cells and a negative correlation with plasma cells. In atopic dermatitis, these three genes exhibited a positive correlation with activated memory CD4+ T cells and a negative correlation with regulatory T cells, activated NK cells, and neutrophils.

Figure 6: Immunoinfiltration analysis using the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm. Note: A-E) Proportions of 22 immune cells in each sample of eczema or atopic dermatitis from GSE57225 and GSE120721 datasets; B-F) Histograms depicting the distribution of immune cell content in GSE57225 and GSE120721 datasets; C-G) Correlations among immune cells in GSE57225 and SE120721 datasets; D-H) Correlation analysis between the expression of three key genes and immune cell content in GSE57225 and GSE120721 datasets.
Simultaneously, this study utilized the ssGSEA algorithm to calculate the relative abundance of various immune cell types in eczema and atopic dermatitis, aiming to obtain more comprehensive results. Display the proportions of 24 immune cells in each eczema or atopic dermatitis sample from the GSE57225 and GSE120721 datasets. Histograms of immune cell content in GSE57225 and GSE120721 are shown in (Figures 7A-7H). In both datasets, the eczema and atopic dermatitis groups exhibited higher proportions of activated dendritic cells, dendritic cells, NK CD56dim cells, T helper cells, Th1 cells, and Th2 cells compared to the normal group. Illustrate the correlations between immune cells in the GSE57225 and GSE120721 datasets. In eczema and atopic dermatitis, activated dendritic cells showed positive correlations with dendritic cells, immature dendritic cells, neutrophils, NK CD56dim cells, T cells, T helper cells, Th1 cells, and Th2 cells, while exhibiting negative correlations with NK cells. Subsequently, Pearson correlation coefficients revealed relationships between immune cell abundance and the expression of key genes. The results indicated that, in eczema and atopic dermatitis, the three key genes showed positive correlations with activated dendritic cells, cytotoxic cells, dendritic cells, neutrophils, NK CD56dim cells, T cells, and plasmacytoid dendritic cells. Additionally, in eczema, the three genes exhibited a positive correlation with Th2 cells, while in atopic dermatitis, GZMB and IRF7 showed a positive correlation with Th2 cells.
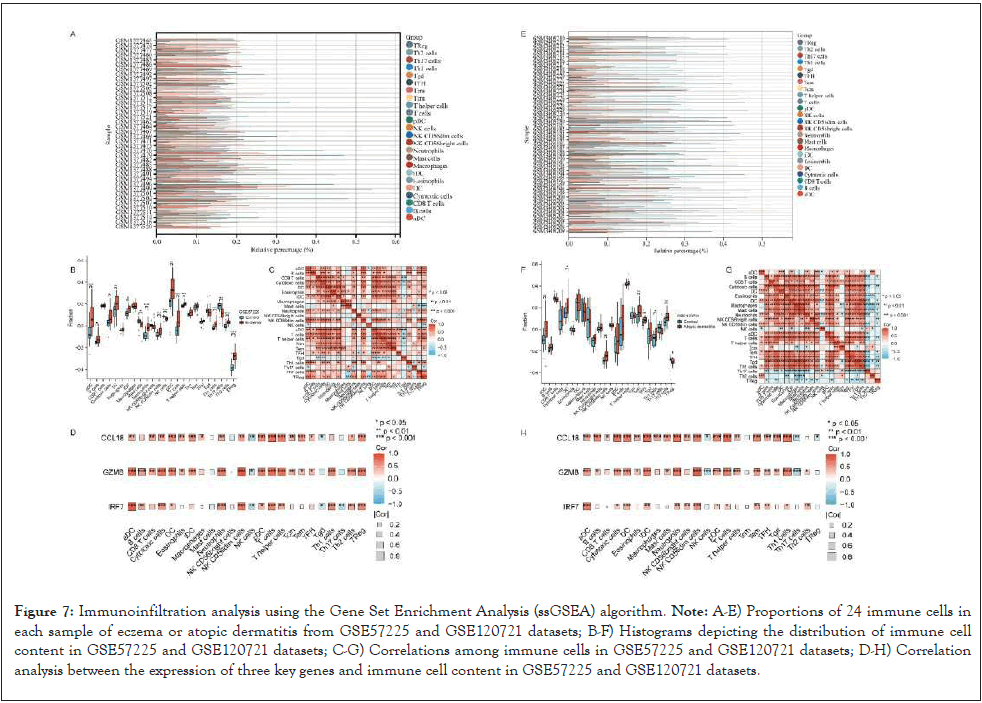
Figure 7: Immunoinfiltration analysis using the Gene Set Enrichment Analysis (ssGSEA) algorithm. Note: A-E) Proportions of 24 immune cells in each sample of eczema or atopic dermatitis from GSE57225 and GSE120721 datasets; B-F) Histograms depicting the distribution of immune cell content in GSE57225 and GSE120721 datasets; C-G) Correlations among immune cells in GSE57225 and GSE120721 datasets; D-H) Correlation analysis between the expression of three key genes and immune cell content in GSE57225 and GSE120721 datasets.
Construction of mRNA-miRNA-lncRNA regulatory networks associated with eczema and atopic dermatitis
Using TargetScan 8.0, miRWalk, and Tarbase v8 databases, miRNA prediction was performed for CCL18, GZMB, and IRF7. Illustrates the network relationships between these three key genes and miRNAs. Subsequently, differential analyses were conducted on GSE175438 and GSE168694. To enhance prediction accuracy, the miRNAs predicted for the three genes were intersected with the upregulated miRNAs in the GSE175438 dataset, obtaining a more consistent set of miRNAs. Then, using the miEAA database, an enrichment analysis was carried out on the obtained miRNA set, revealing their involvement in various pathways such as fatty acid biosynthesis, leukocyte transendothelial migration, chemokine signaling pathway, PI3K-Akt signaling pathway, JAK-STAT signaling pathway, B cell receptor signaling pathway, MAPK signaling pathway, Th1 and Th2 cell differentiation, Tumor Necrosis Factor (TNF) signaling pathway, cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway, NF-κB signaling pathway, T cell receptor signaling pathway, and Th17 cell differentiation (Figures 8A-8H).
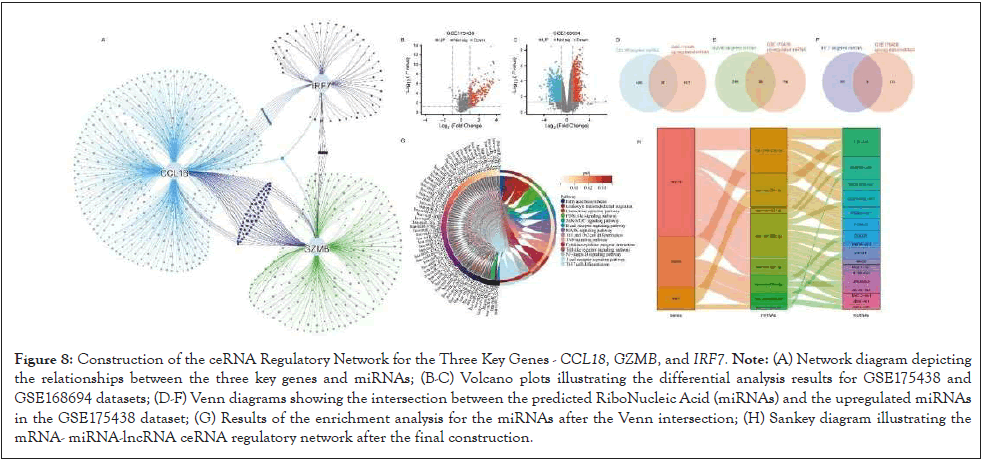
Figure 8: Construction of the ceRNA Regulatory Network for the Three Key Genes - CCL18, GZMB, and IRF7. Note: (A) Network diagram depicting the relationships between the three key genes and miRNAs; (B-C) Volcano plots illustrating the differential analysis results for GSE175438 and GSE168694 datasets; (D-F) Venn diagrams showing the intersection between the predicted RiboNucleic Acid (miRNAs) and the upregulated miRNAs in the GSE175438 dataset; (G) Results of the enrichment analysis for the miRNAs after the Venn intersection; (H) Sankey diagram illustrating the mRNA- miRNA-lncRNA ceRNA regulatory network after the final construction.
Within these pathways, it was discovered that 11 miRNAs were involved in all of them. Through the miRNet 2.0 database, lncRNA prediction was performed for these 11 miRNAs, and eight miRNAs were predicted to target lncRNAs. To enhance accuracy, the predicted lncRNAs were intersected with the upregulated lncRNAs in the GSE168694 dataset, completing the construction of the final mRNA-miRNA- lncRNA ceRNA regulatory network.
Single-Cell transcriptomic landscape and immunohistochemical expression patterns of CCL18, IRF7, and GZMB in atopic dermatitis and eczema
To deepen our understanding of atopic dermatitis and eczema, we explored the ADGSE197023 dataset, enabling us to assess the expression of the three genes of interest at single-cell resolution. Initially, the cell population was classified into six clusters. Subsequently, these clusters were categorized into three distinct cellular populations based on the expression of marker genes: fibroblasts, keratinocytes, and suprabasal keratinocytes. We found that CCL18 and IRF7 were expressed in all three cell types, while GZMB was primarily expressed in fibroblasts. Further, we conducted pseudotime analysis using Monocle 2, which plotted a tree-like developmental trajectory, illustrating the different cellular developmental states. The developmental trajectories of fibroblasts and suprabasal keratinocytes were similar, being present in the early and middle stages of the trajectory and decreasing in the later stages. Keratinocytes were present in the early stages of the trajectory, however, as the trajectory progressed towards the middle stages, this cell type diminished, only to increase again in the later stages (Figures 9A-9K). To validate the findings from single- cell analysis, we performed immunohistochemical experiments to assess the protein expression levels of CCL18, IRF7, and GZMB in control (NC), eczema, and AD tissues. The results indicated that, compared to the control group, the protein expression levels of CCL18, IRF7, and GZMB were all highly expressed in both eczema and AD tissues.

Figure 9: Single-Cell analysis and validation in Atopic dermatitis and eczema. Note: (A) Cell markers for clusters’annotation; (B) Cell clusters for GSE197023 of AD samples; (C) Uniform Manifold Approximation and Projection (UMAP) visualization of all cells in 6 clusters; (D-F) Expression of CCL18, IRF7 and GZMB in 3 subpopulations of cells; (G) Monocle-based pseudotime trajectory colored by pseudotime; (H) Monocle 2 Pseudotime Trajectory Analysis of Atopic Dermatitis Cells; (I-K) Immunohistochemical analysis of CCL18, IRF7, and GZMB expression in atopic dermatitis and eczema; (G) Monocle-based pseudotime trajectory colored by pseudotime; (H) Monocle 2 Pseudotime Trajectory Analysis of Atopic Dermatitis Cells; (I-K) Immunohistochemical analysis of CCL18, IRF7 and GZMB expression in atopic dermatitis and eczema.
This study employed an integrative approach combining Weighted Gene Co-expression Network Analysis (WGCNA), machine learning, and bioinformatics methods to comprehensively dissect the comorbid mechanisms of eczema and atopic dermatitis. The goal was to identify potential therapeutic targets and provide a deeper understanding of these two skin conditions. Selecting datasets from the Gene Expression Omnibus (GEO), including GSE6012, GSE14550, GSE32924, and GSE120721, we conducted Weighted Gene Co-expression Network Analysis (WGCNA) and Differential Expression Genes (DEGs) analysis. By intersecting the results of these analyses, we successfully identified 22 genes that were co-expressed in both eczema and atopic dermatitis. These genes underwent Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, disease prediction, and Protein-Protein Interaction (PPI) analysis. Random Forest in machine learning and the cytohubba plugin in Cytoscape software further filtered out three key hub genes: CCL18, GZMB, and IRF7. The application of CIBERSORT and ssGSEA for immune infiltration analysis revealed the close relationship of these three genes with immune cells. A mRNA- miRNA-lncRNA ceRNA regulatory network was established. Enrichment analysis results indicated that these 22 genes are involved in various biological processes such as cytokine-mediated signaling pathways, leukocyte migration, leukocyte chemotaxis, neutrophil chemotaxis, and humoral immune responses, showing associations with multiple skin-related diseases, including atopic dermatitis. Consistency between machine learning and network analysis confirmed the essential role of CCL18, GZMB, and IRF7. Further immune infiltration analysis revealed the associations of these three genes with different immune cell types.
The comprehensive findings of this study suggest that CCL18, GZMB, and IRF7 may serve as potential biomarkers for eczema and atopic dermatitis, holding significant clinical promise. CCL18, belonging to the chemokine family, has been reported to exhibit highly induced expression in the lesional skin of atopic dermatitis patients [11,12]. Studies, including research by Satoru Sanada et al., have confirmed the involvement of CCL18 in the pathogenesis of diseases such as cystic fibrosis, rheumatoid arthritis, and atopic dermatitis [13]. GZMB, a serine protease with cytotoxic and immune-regulatory functions, has been found to have elevated levels in the plasma of atopic dermatitis patients. Moreover, in the skin lesions of atopic dermatitis patients, the expression of GZMB in CD4+ and CD8+ T cells is significantly higher than in healthy skin. The mRNA expression of GZMB is notably increased in Th2- polarized CD4+ T cells and IL-4-activated CD4+ T cells, with histamine and its H2 receptor agonist significantly enhancing GZMB levels [14]. Interferon Regulatory Factor 7 (IRF7), a member of the interferon regulatory transcription factor family, plays a crucial role in various biological processes such as inflammation, apoptosis, and immune response. Studies indicate that IRF7 is upregulated in type 2 lymphocytes during allergic inflammatory processes, and its deficiency leads to attenuation [15]. Specifically, research by Cohen. reported the regulatory role of IRF7 in the phenotypic transition from pro-inflammatory macrophages to anti-inflammatory macrophages (M1 to M2), a process negatively regulated by the transforming growth factor-β1 pathway [16]. Chenyang et al., through external dataset validation and immunohistochemical analysis, identified five central genes, including IRF7, as potential diagnostic and therapeutic biomarkers for atopic dermatitis.
Through immune infiltration analysis using CIBERSORT and ssGSEA algorithms, we delved into the immune cell landscape in eczema and atopic dermatitis tissues. In eczema, a significant increase in initial CD4+ T cells, activated memory CD4+ T cells, and M1 macrophages was observed, while the content of plasma cells and resting mast cells was lower. In contrast, in atopic dermatitis, the proportion of activated memory CD4+ T cells was higher, while regulatory T cells, activated NK cells, and resting mast cells were relatively lower. These differences underscore the immunological heterogeneity at the cellular level in eczema and atopic dermatitis.
Regarding the association between key genes and immune cells, we found that in eczema, the three key genes are positively correlated with activated memory CD4+ T cells and negatively correlated with resting mast cells. In atopic dermatitis, these three genes are positively correlated with activated memory CD4+ T cells and negatively correlated with regulatory T cells, activated NK cells, and neutrophils. Previous studies have indicated that inflammatory skin diseases, such as psoriasis, atopic dermatitis, and contact dermatitis, involve the recruitment of T cells [17]. In patients with atopic dermatitis, CCL18 binds to skin-homing CLA+ memory T cells, triggering the migration of human memory T cells in vivo. Further analysis using the ssGSEA algorithm showed a significant increase in the proportions of activated dendritic cells, dendritic cells, NK CD56dim cells, T helper cells, Th1 cells, and Th2 cells in eczema and atopic dermatitis. Literature studies also confirm the existence of pathogenic Th2 cell subsets in the peripheral blood of patients with atopic dermatitis [18], and the excessive activation of Th2 cells often manifests as allergic symptoms such as rhinitis, atopic dermatitis, and asthma [19], emphasizing the key role of Th2 cells in the pathological development of atopic dermatitis [20]. In eczema, the three genes are positively correlated with Th2 cells, while in atopic dermatitis, GZMB and IRF7 are positively correlated with Th2 cells. These findings further support the close association between the three key genes and eczema and atopic dermatitis.
The incorporation of single-cell transcriptomic analysis in our study offers a refined perspective on the cellular heterogeneity within eczema and atopic dermatitis lesions. The distinct cellular clusters identified, along with their specific expression patterns for CCL18, IRF7, and GZMB, provide a foundation for understanding the cellular dialogue in these conditions. The pseudotime trajectory analysis elucidates the dynamic changes in cellular populations, suggesting a temporal sequence in disease progression. The immunohistochemical validation of these genes strengthens the link between molecular signatures and observable protein expression in the skin lesions. The increased expression of CCL18, IRF7, and GZMB in eczema and AD tissues, as compared to controls, underscores their potential as diagnostic markers and therapeutic targets. Our findings suggest that the cellular and molecular alterations captured through single-cell analysis and corroborated by immunohistochemistry are pivotal to the pathogenesis of eczema and AD. These genes may represent critical nodes in the inflammatory networks driving these diseases, offering targets for intervention. The trajectory analysis hints at a temporal regulation of key cellular processes, which could inform stage- specific therapeutic strategies. Targeting these genes at particular stages of the disease may prove more efficacious than broad immunosuppression. While our study provides valuable insights, future research should focus on the functional implications of the observed gene expression patterns. In-depth cellular studies are needed to determine the exact roles of CCL18, IRF7, and GZMB in disease pathogenesis.
This study was conducted in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments. It was approved by the Ethics Committee of the Women and Children's Hospital of Hunan, Changsha, China (Approval Number: 2024-13).
The authors have no conflict of interest.
We acknowledge the miRNet 2.0, GEO, STRING, miEAA, TargetScan 8.0, miRWalk, Tarbase v8, CIBERSORTx and Metascape databases for free use.
Youcai Yan conceived and designed the experiment, supervised the project, critically reviewed the manuscript, edited it for clarity and coherence, and finally approved the submitted version. Fang Lin contributed to manuscript drafting and bioinformatics analysis, while Cong Lin conducted some of the bioinformatics analyses.
The datasets supporting the conclusions of this study are available in the Gene Expression Omnibus (GEO) repository, which is a public database. The following are the accession numbers for the datasets used in this research: GSE6012, GSE14550, GSE32924, and GSE120721. The raw data can be accessed through the GEO database without any special permission or restrictions. All other data generated or analyzed during this study are included in this published article. Additional information and reasonable requests for data or materials can be directed to the author, Fang Yang (Email: 18373701128@163.com).
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
[CrossRef] [Google Scholar] [PubMed]
Citation: Yang F, Lin C, Yan Y (2024). Deciphering Molecular Underpinnings and Therapeutic Avenues for Eczema and Atopic Dermatitis through Integrated Omics and Immunohistochemical Analysis. Chemo Open Access. 12:225
Received: 18-Nov-2024, Manuscript No. CMT-24-34560; Editor assigned: 20-Nov-2024, Pre QC No. CMT-24-34560 (PQ); Reviewed: 04-Dec-2024, QC No. CMT-24-34560; Revised: 11-Dec-2024, Manuscript No. CMT-24-34560 (R); Published: 19-Dec-2024 , DOI: 10.35248/2167-7700.24.12.225
Copyright: © 2024 Yang F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited