Journal of Theoretical & Computational Science
Open Access
ISSN: 2376-130X
ISSN: 2376-130X
Research Article - (2023)Volume 9, Issue 4
As hemp-based cannabinoids are continuously gaining popularity, synthesis and extraction methods for these compounds are ever-changing. Within the cannabinoid market, hydrogenated derivatives are also gaining popularity at an accelerated rate, with the need for in-depth analysis of these compounds pertinent to increasing the knowledge of cannabis chemistry. Our lab used Schrodinger to dock natural and synthetic cannabinoids in various CB 1 and CB 2 receptors, PPAR-γ, PAK1 and GPR119 complex including several enzymes, to evaluate the interacting residues within the known binding pockets, comprising the computation of binding energies, predicting ADME characteristics and evaluating P450 sites of metabolism. The purpose of identifying active residues, sites of metabolism and ADME characteristics for 40 various cannabinoids is to provide guidance in computer-aided drug design and rationalization in designing and synthesis of analogs.
In silico; Computational chemistry; GPCR; Cannabinoids; CB 1; CB 2; ADME
ADMET: Adsorption, Distribution, Metabolism, Excretion and Toxicity; Arg: Arginine; Asp: Aspartic Acid; CBC: Cannabichromene; CBCA: Cannabichromenic Acid; CBD: Cannabidiol; CBDA: Cannabidiolic Acid; CBG: Cannabigerol; CBGA: Cannabigerolic Acid; CBR: Cannabinoid Receptor; Cys: Cystine; DFT: Density Funtional Theory; GC-MS: Gas Chromatography-Mass Spectrometry; Glu: Glutamic Acid; H4CBD: Hexahydrocannabidiol; H-bonding: Hydrogen bonding; HHC: Hexahydrocannabinol; HHCV: Hexahydrocannabivarin; His: Histidine; IC50: Half-maximal Inhibitory Concentration; Ile: Isoleucine; Leu: Leucine; Met: Methionine; MTT: 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; NAM: Negative Allosteric Modulator; PAM: Positive Allosteric Modulator; PDB: Protein Database; Phe: Phenylalanine; RCSB: Research Collaboratory for Structural Bioinformatics; RMSD: Root Mean Square Deviation; Ser: Serine; THC: Tetrahydrocannabinol; THCA: Tetrahydrocannabinolic Acid; THCBC: Hydrogenated CBC; THCBG: Hydrogenated CBG; THCV: Tetrahydrocannabivarin; Thr: Threonine; TMSCl: Chlorotrimethylsilane; Trp: Tryptophan; Tyr: Tyrosine; Val: Valine; CB 1: Cannabinoid receptor 1; CB 2: Cannabinoid receptor 2; PPAR- γ: Peroxisome Proliferator-Activated Receptor Gamma; PAK1: Serine/Threonine Protein Kinase; SOM: Sites of Metabolism; Caco-2: Colorectal Adenocarcinoma-2; MMGBSA: Molecular Mechanics Generalized Born Surface Area; CBN: Cannabinol
Cannabidiol (CBD) and Tetrahydrocannabinol (THC) are the major cannabinoids biosynthesized by Cannabis sativa, yet there are cannabinoids to be elucidated within the hundreds of compounds that are naturally biosynthesized. The elucidation of these cannabinoid compounds also promotes the creation of semi- to fully synthetic cannabinoids, to mimic the natural scaffolds and their effects. Recently, a wave of semi-synthetic cannabinoids is beginning to appear in smoke shops and dispensaries both nationally and internationally [1-3]. A growing trend of unqualified personnel performing synthetic chemistry is of concern due to the potential for hazardous byproducts that might remain despite purification [4]. Since their identification in the 1940s, hydrogenated cannabinoids have reappeared within consumer and retail markets as alternative solutions to overtightening regulations and bills in place to limit and restrict cannabinoids derived from hemp or marijuana [5-10]. Cannabigerol (CBG) and Cannabichromene (CBC), are considered minor constituents within the cannabinoid biome produced by C. sativa. CBG is also considered an important precursor to the transformation to CBC, the formation of CBD and THC, through a known biosynthetic pathway (Figure 1). Harvey, et al., [11,12] reported metabolites of Tetrahydrocannabigerol (THCBG) and Tetrahydrocannabichromene (THCBC) using TMSCl derivatization and GC-MS. ElSohly, et al., [13] tested the saturated cannabinoids identifying antimicrobial and antifungal properties which demonstrate that the saturation of CBG and CBC olefins led to an increase in the anti-microbial and anti-fungal characteristics [14].
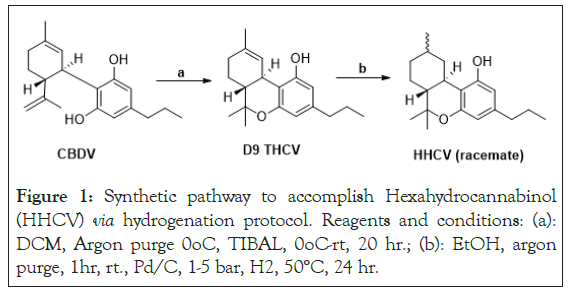
Figure 1: Synthetic pathway to accomplish Hexahydrocannabinol (HHCV) via hydrogenation protocol. Reagents and conditions: (a): DCM, Argon purge 0oC, TIBAL, 0oC-rt, 20 hr.; (b): EtOH, argon purge, 1hr, rt., Pd/C, 1-5 bar, H2, 50°C, 24 hr.
Tesfatsion, et al., [15] demonstrated that saturation of Tetrahydrocannabivarin (THCV) to yield Hexahydrocannabivarin (HHCV), as shown in Figure 1, improved IC50 values in PANC-1 MTT assays.
Novel Hexahydrocannabinol (HHC) analogs have also shown promise as anticancer agents from cell studies to xenograft models [15,16-18]. Saturated cannabinoids in the literature have shown promise with medicinal properties compared to their unsaturated counterparts [5]. Lovering, et al., [19] discussed an increase in saturation or fraction sp3 and the presence of chiral centers within molecules leads to an increase in the ability for discovery drugs to reach commercialization. In previous work, we reported that IC50 values of HHCV and THCV in-vitro screening using MTT assays on the proliferation of PANC-1 pancreatic cell line are 5.5 µM and 14.7 µM, respectively. Also, saturated cannabinoids CCL104 and CCL105 exhibited 1.06 µM and 2.55 µM as IC50 values on the same pancreatic cell line. On the other hand, we demonstrated that R-HHC is more active than S-HHC in PANC-1 pancreatic cancer cells showing 10.3 µM and 18.9 µM as IC50 values, respectively [15].
The question of where these hydrogenated compounds bind, how they are metabolized and the nature of their toxicity profiles remain unreported. Using Schrodinger, our group has performed in-silico experiments using QikProp, LigPrep, Jaguar, ADMET, Glide, Epik, Desmond, Phase, Protein preparation wizard and sitemap to identify binding interactions and predicted binding scores, predicted ADME, predicted p450 metabolism and metabolites for a series of saturated and non-saturated cannabinoids to compare the difference among these two groups of cannabinoids.
In literature, Cannabinoid receptor 1 (CB 1) and Cannabinoid receptor 2 (CB 2) belong within the family of GPCRs and are known to bind with cannabinoids enacting physiological and psychological effects [20,21]. The use of these receptors focuses on treating diseases, using cannabinoids and similar cannabimimetic compounds, enact agonistic or antagonistic effects, such as anticancer or anti-inflammatory responses when the bound receptors are activated or deactivated [21]. Other receptors were selected due to the similarity of the GPCR family or in relation to the diseases the receptors are implicated in to determine the effects of whether classical cannabinoids or hydrogenated analogs bind within their domains. Proteins and enzymes of interest were chosen from the RCSB protein data bank, which includes Peroxisome Proliferator-Activated Receptor Gamma (PPAR- γ), Serine/Threonine Protein Kinase (PAK1), CB 1 receptors, CB 2 receptors and GPR119. The reasoning for selected receptors and kinases is due to their implication in disease as well as actively studied mechanisms of cannabinoid binding that include these proteins. Each of the proteins was crystallized with either an agonist or antagonist and was used as references in binding cannabinoid ligands to the binding domain.
PPAR-γ is a type II nuclear receptor that functions as a transcription factor [22]. Many agents directly bind and activate PPAR-γ, some include fatty acids and cannabinoids. Activation of PPAR-γ might be responsible in the inhibition of breast, gastric, lung and prostate cancer cell lines [22,23]. PAK1 regulates cytoskeleton remodeling, phenotypic signaling and gene expression. PAK1 is associated with a wide variety of cellular processes such as directional motility, invasion, metastasis, growth, cell cycle progression and angiogenesis [24].
PAK1-signaling-dependent cellular functions regulate both physiologic and disease processes, including cancer, due to overexpression in human cancer [24]. Nikfarjam, et al., [25] demonstrated CBD and THC practice their inhibitory effects on pancreatic cancer via a PAK1-dependent pathway, indicating that CBD and THC cancel the kras protein-activated pathway by affecting PAK1.
GPR119 a novel cannabinoid receptor, is found within the pancreas and intestinal tract with implication on affecting incretin and insulin hormone secretions, with novel drug discovery using this receptor to treat obesity and diabetes [26].
Chosen CB 1 receptors from RCSB with no conformational changes were used to dock the ligands, compared to selected CB 1 proteins that contain a Negative Allosteric Modulator (NAM) bound to it enacting conformational change. Both types were used to compare the differing residue interactions that might occur. Conformationally changed proteins may enact different effects, which is why they were chosen to potentially observe differing residue interactions [27].
CB 2 receptors were selected with a similar parameter to CB 1 receptors, identifying non-conformationally changed and conformationally changed proteins and their differing residue interactions to bound cannabinoid ligands. The GPR119 complex in the GPCR family is thought to be a part of the mechanism in which cannabinoids express their effects.
The compounds that were bound within the receptors exhibited cation-π stacking, π - π stacking and H-bonding primarily. The interactions that were seen are highly important biological connections that strengthen ligand binding energies within the receptors. The cation- π interaction is shown to increase binding energy by ~2.6 kcal/mol and π - π stacking additionally is seen to contribute to ligand stability within the receptor binding pocket [28,29]. Some of the π – π stacking conformations include sandwich, T-shaped and parallel displaced, due to the ligand conformation. H-bonding was also seen, with the solvent effect and interaction with various water molecules, amino acid residues and intercalation of water molecules to amino acids, the bonding kcal can vary from 1-40 kcal/mol.
Cannabinoid agonists that activate the Cannabinoid Receptors (CBR) initiate pathways that can lead to inhibition or activation ultimately leading to the blocking of cell cycle, proliferation, cell death, angiogenesis, metastases and cellular transition. Derived proteins as mentioned were pulled from the activated/deactivated pathways which correlate disease genesis or progression (Figure 2) [30-40].
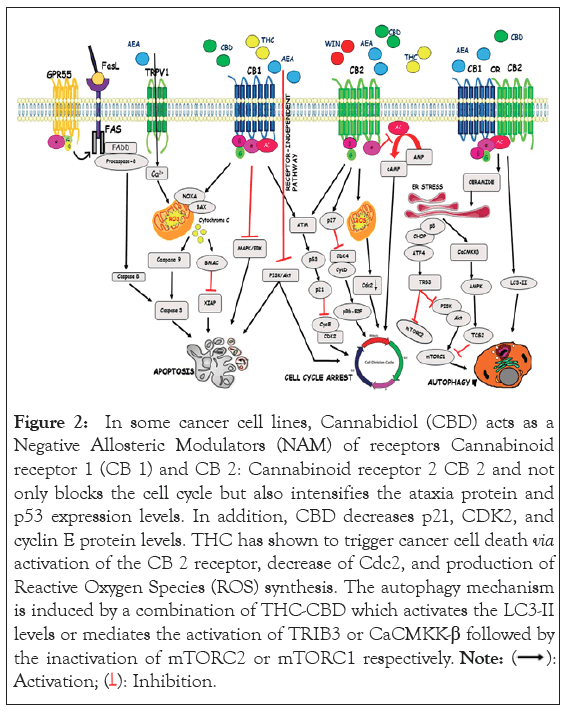
Figure 2: In some cancer cell lines, Cannabidiol (CBD) acts as a Negative Allosteric Modulators (NAM) of receptors Cannabinoid receptor 1 (CB 1) and CB 2: Cannabinoid receptor 2 CB 2 and not only blocks the cell cycle but also intensifies the ataxia protein and p53 expression levels. In addition, CBD decreases p21, CDK2, and cyclin E protein levels. THC has shown to trigger cancer cell death via activation of the CB 2 receptor, decrease of Cdc2, and production of Reactive Oxygen Species (ROS) synthesis. The autophagy mechanism is induced by a combination of THC-CBD which activates the LC3-II levels or mediates the activation of TRIB3 or CaCMKK-β followed by the inactivation of mTORC2 or mTORC1 respectively. 
A proteins and ligands preparation
All Molecular docking experiments were achieved on Cybertron PC CLX 13th Gen Intel(R) Core(TM) i9-13900KF @ 3.00 GHz comprising 24 computing cores. Schrödinger Release 2023-3: Glide software was used as the docking program [26]. Crystal structures of CB 1, CB 2, GPR119, PAK1 and PPAR-γ were retrieved from the RCSB protein data bank. CB 1 [(PDB: 7V3Z), (PDB: 5U09), (PDB: 6KQI)]. CB 2 [(PDB: 5ZTY), (PDB:6PT0), (PDB: 6KPC)]. GPR119 [(PDB: 7WCM)]. PAK1 [(PDB: 5DFP)]. PPAR-γ [(PDB: 2P4Y)].
The proteins were prepared using a protein preparation workflow tool on Schrodinger Protein Preparation Wizard [27]. The external water molecules and ions were removed. Polar Hydrogens were added. Missing side chains were filled using Epic and PROPKA. Het states were generated at pH 7.4 (+/- 2.0). Heavy atoms converged to RMSD 0.30Å. 3D structures of cannabinoids and hydrogenated cannabinoids were established in 2D sketcher which was then exported as an SDF file and imported and prepared using LigPrep, to form 3D conformers, including the various 3D chiral conformations. All structures underwent geometrical optimization using release 2023-3: Jaguar software using Density Functional Theory (DFT) calculation with B3LYP/6-31G as the basis set for the calculation to afford the minimized energy chemical structures. The structures were then docked using release 2023-3: Glide software from Schrödinger.
In silico molecular docking
The grid parameter was generated covering the CB 1 pockets for (PDB:7V3Z) [-42.91, -163.58, 306.7], (PDB:5U09) [126.7,118.85,147.7], (PDB:6KQI) [-25.98, -8.77, 40.11] for x, y and z coordinates. The ligand diameter midpoint box follows a 10Å × 10Å × 10Å x, y, z dimension. The grid parameter was generated covering the CB 2 pockets for (PDB:5ZTY) [9.09, -0.17, -55.72], (PDB:6PT0) [98.38, 109.56, 123.8], (PDB:6KPC) [10.52, 1.26, -45.17] for x, y, z coordinates. The Ligand diameter midpoint box follows a 10Å × 10Å × 10Å x, y, z dimension. The grid parameter was generated covering the GPR119 pocket (PDB:7WCM) [126.7, 118.85, 147.7] for x, y, z coordinates. The ligand diameter midpoint box follows a 10Å × 10Å × 10Å x, y, z dimension. The grid parameter was generated covering the PAK 1 pocket (PDB:5DFP) [13.58, 34.37, -15.61] for x, y, z coordinates. The ligand diameter midpoint box follows a 10Å × 10Å × 10Å x, y, z dimension. The grid parameter was generated covering the PPAR-γ pocket (PDB:2P4Y) [35.4, -21.89, 39.56_B] for x, y, z coordinates. The ligand diameter midpoint box follows a 10Å × 10Å × 10Å x, y, z dimension.
The minimized energy structures were received using Jaguar software, Density Functional Theory (DFT) calculation with B3LYP/6-31G as the basis set for the calculation and prepared proteins using the protein preparation workflow tool on the Maestro 12.5 interface of Schrödinger protein preparation wizard [27]. Prime MM–GBSA (MMGBSA dG Bind (NS) and MMGBSA dG Bind) energy was calculated and displayed in Supplementary Table 1. MM/GBSA calculations were accomplished to esteem the relative binding energies of cannabinoids to the receptors.
Prediction of ADMET properties
The Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties of the 40 cannabinoids were performed using QikProp version 4.4 integrated into maestro (Schrodinger, LLC, New York, 2015) which predicts the widest variety of pharmaceutically relevant properties: predicted aqueous solubility (QPlogS), predicted IC50 value for blockage of Human Ether-a-go-go Related Gene (HERG) Potassium+ (K+) channels (QPlogHERG), predicted apparent Caco-2 cell permeability (QPPCaco) Colorectal Adenocarcinoma-2 (Caco-2) cells are a model for the gut-blood barrier), predicted brain/blood partition coefficient (QPlogBB) and % human oral absorption (Predicted human oral absorption in gastrointestinal tract on 0 to 100% scale). The calculated physicochemical descriptors are displayed in Supplementary Table 2. QikProp bases its predictions on the full 3D molecular structure and the global minimum energy conformer of each compound was used as input for ADMET properties.
Hypothesized P450 sites of metabolism
Schrodinger P450 site of metabolism software was used to perform calculations. Cytochrome (CYP) isoform (intrinsic reactivity) function was used to determine possible Sites of Metabolism (SOM).
Molecular docking of cannabinoids
We used a virtual screen of 40 identified natural and synthetic cannabinoids and their diastereomers to explore the binding interaction between cannabinoids and PPAR-γ (2P4Y binding domain complexed with C03: Figure 3), PAK1 (5DFP in complex with an inhibitor compound FRAX1036: Figure 3), CB 1 receptors (5U09 bound to an inverse agonist 7DY, 6KQI bound to an agonist modulator CP55940 and 7V3Z bound to CP55940: Figure 3), CB 2 receptors (5ZTY bound to high-affinity synthetic antagonist AM10257, 6PT0 in complex with agonist WIN 55,212-2 and 6KPC with E3R an agonist bound GPCR: Figure 3) and GPR119 complex (7WCM with an agonist MBX-2982: Figure 3) We selected the cannabinoids to be docked considering three main structural components: the aliphatic side chain (C1-C7 and adamantyl) at the meta-position of the phenol in the aromatic ring, saturated or not saturated ring of the terpene moiety and monocyclic, bicyclic or tricyclic cannabinoids. The compounds that were screened included CBD, THC, CBC, CBG and CBN with different substituents in the side chain and their hydrogenated analogs: H4CBD, HHC, THCBC and THCBG (Figure 4).
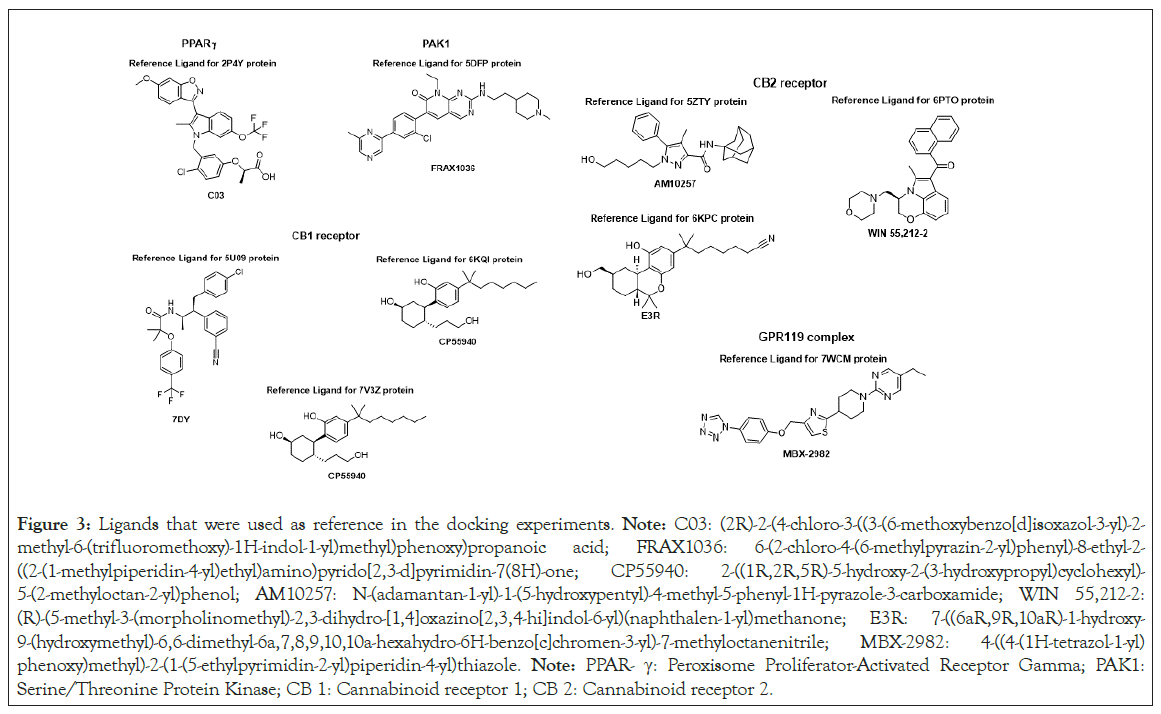
Figure 3: Ligands that were used as reference in the docking experiments. Note: C03: (2R)-2-(4-chloro-3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-methyl-6-(trifluoromethoxy)-1H-indol-1-yl)methyl)phenoxy)propanoic acid; FRAX1036: 6-(2-chloro-4-(6-methylpyrazin-2-yl)phenyl)-8-ethyl-2-((2-(1-methylpiperidin-4-yl)ethyl)amino)pyrido[2,3-d]pyrimidin-7(8H)-one; CP55940: 2-((1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl)-5-(2-methyloctan-2-yl)phenol; AM10257: N-(adamantan-1-yl)-1-(5-hydroxypentyl)-4-methyl-5-phenyl-1H-pyrazole-3-carboxamide; WIN 55,212-2: (R)-(5-methyl-3-(morpholinomethyl)-2,3-dihydro-[1,4]oxazino[2,3,4-hi]indol-6-yl)(naphthalen-1-yl)methanone; E3R: 7-((6aR,9R,10aR)-1-hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl)-7-methyloctanenitrile; MBX-2982: 4-((4-(1H-tetrazol-1-yl)phenoxy)methyl)-2-(1-(5-ethylpyrimidin-2-yl)piperidin-4-yl)thiazole. Note: PPAR- γ: Peroxisome Proliferator-Activated Receptor Gamma; PAK1: Serine/Threonine Protein Kinase; CB 1: Cannabinoid receptor 1; CB 2: Cannabinoid receptor 2.
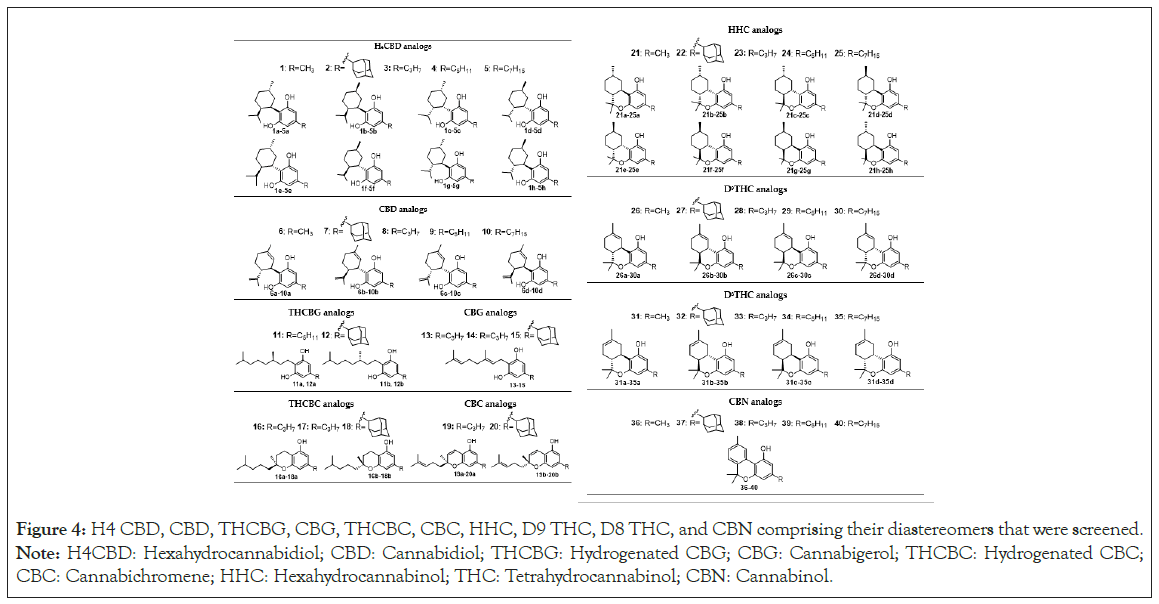
Figure 4: H4 CBD, CBD, THCBG, CBG, THCBC, CBC, HHC, D9 THC, D8 THC, and CBN comprising their diastereomers that were screened. Note: H4CBD: Hexahydrocannabidiol; CBD: Cannabidiol; THCBG: Hydrogenated CBG; CBG: Cannabigerol; THCBC: Hydrogenated CBC; CBC: Cannabichromene; HHC: Hexahydrocannabinol; THC: Tetrahydrocannabinol; CBN: Cannabinol.
Howlett, et al., [41] demonstrated that D9 THC (compound 29) shows partial agonist activity at CB 1 (Ki=10 nM) and CB 2 receptors (Ki=24 nM). Later Tham, et al., [42] reported that CBD (compound 9) displays a partial agonism and orthosteric site binding with CB 2 receptor with Ki>10000. Finally, Chung, et al., [43] revealed the affinity of D9 THCP (compound 30) to both CB 1 and CB 2, with a Ki of 1.2 and 6.2 nM, respectively which are higher than D9 THC. THCP binds to CB 1 receptor in an L-shaped pose with the chromene ring moiety occupying the hydrophobic pocket. However, there is no systematic study of the interactions and relative free energies of different cannabinoids with CB 1 and CB 2 receptors to demonstrate the influence of the side chain, the double bond in the terpene ring and the bicyclic and tricyclic system in the binding pocket with these receptors.
Using Jaguar to perform minimizations and calculate DFT for given scaffolds, the then minimized scaffolds were docked within the various proteins that were prepared using the Schrödinger protein preparation workflow. The relative binding energy and docking scores of all docked cannabinoids were calculated to classify the intensity of protein-ligand interactions (Supplementary Table 1-9).
The docking results showed that compounds, 10, 30, 32, 35, 37-39 were not successfully docked into the CB 1, CB 2, GPR119, PAK1 and PPAR-γ models. The most favorable pose for each cannabinoid was chosen and analyzed. The docking scores ranged from -3.031 kcal/mol to -10.949 kcal/mol and they are recorded in Supplementary Table 1-11. Compound 14 presented the most promising docking score of -10.949 kcal/mol with 6KPC protein (CB 2 receptor) and compound 17a showed the least promising docking score of -3.282 kcal/mol with 5DFP (PAK1 receptor). The relative binding energies were determined by the prime MM-GBSA module and extended from -86.054 kcal/mol (18b:7V3Z complex) to -15.232 kcal/mol (1e:5U09 complex) for cannabinoids (Supplementary Table 1-9).
Supplementary Table 1-9 shows the interacting residues and interaction types of cannabinoids that were coupled. The common motifs of the docked cannabinoids were π-cation, H-bonding and π - π stacking. All the residues were within 4Å of the interacting moiety.
PPAR-γ (2P4Y): Supplementary Table 3 show the cannabinoids and their interactions with the PPAR-γ (2P4Y model). CBG-5C (14) was demonstrated to exhibit the greatest favorable docking score of -8.241 kcal/mol and CBG-3C (13) was proven to display the least promising docking score of -4.772 kcal/mol (Supplementary Table 10). CBG-5C (14) has H-bonding with Leu340 residue and CBG-3C (13) has hydrogen bond with H2O. Cannabinoids (CBD-5C:9c, HHC-1C:21f, HHC-3C:23c) that resulted in high docking scores and relative binding energies ranged between -42.402 kcal/mol and -50.369 kcal/mol (Supplementary Table 3) presented multiple interactions with the 2P4Y protein: H-bond interaction between phenolic hydroxyl groups (from resorcinol moiety) and Leu340, Cys285 and H2O residues. Also, the resorcinol ring exhibited π-cation interaction with Arg288 residue. It is interesting to note that compound 26b (D9 THC-1C) exhibited the strongest relative binding energy complex D9 THC-1C:2P4Y Molecular Mechanics Generalized Born Surface Area (MMGBSA) dG Bind(NS)=59.605 kcal/mol: Supplementary Table 3) with a very good docking score (-7.553 kcal/mol) and was the only cannabinoid that displayed H-bonding interaction with Ser289 (Supplementary Figure 1), which is the major interaction presented by the indole reference ligand ((2R)-2-(4-chloro-3-{[3-(6-methoxy-1,2-benzisoxazol-3-yl)-2-methyl-6-(trifluoromethoxy)-1H-indol-1yl]methyl}phenoxy) propanoic acid). Brunsveld, et al., [44] proved that indazole MRL-871 interacts with PPARγ Ser289 residue via hydrogen bond and plays a key role in the stabilization of the beta-sheet region of the PPARγ receptor.
PAK1 (5DFP): Supplementary Table 1 indicate the interactions of docked cannabinoids with the PAK1 (5DFP) model and the FRAX1036 as reference inhibitor ligand. The docking results showed that 25 cannabinoids out of 40 were successfully docked into the 5DFP protein. The highest docking score corresponds to CBN-7C (40) with -6.309 kcal/mol and the lowest is -3.031 kcal/mol for CBD-Adamantyl (7) as shown in Supplementary Table 1. CBN-7C (40) interacted with Asp393 and H2O forming a conventional hydrogen bond with each of these two residues. Interaction of CBN-7C (40) with 5DFP, are shown in Supplementary Figure 2. The complex of CBN-7C (40):5DFP was found to exhibit the strongest MMGBSA dG Bind (NS) energy with -57.664 kcal/mol (Supplementary Table 3).
The frequent interaction pattern that was observed among the cannabinoids and the residues includes π-cation (Arg299 with phenyl ring from resorcinol moiety) and aromatic hydrogen bond (Thr406, Leu347, Gluc345, Gluc315, Asp393, H2O). The interaction pattern was compared with the reference inhibitor FRAX1036 of the PAK1 crystal structure which showed hydrogen bond interactions with Glu67, Gluc315, H2O, Arg51, Leu99, Asp 106, Asp393 and Thr406.
In addition, compounds H4 CBD-3C (3d), CBD-3C (8b), HHC-1C (21f), HHC-3C (23a), HHC-5C (24b), D8 THC-3C (33d) and D8 THC-5C (34c) revealed two interactions with the amino acid residues showing -5.333 kcal/mol, -5.288 kcal/mol, -5.249 kcal/mol, -5.909 kcal/mol, -5.162 kcal/mol, -5.846 kcal/mol and -5.604 kcal/mol as docking scores, respectively (Supplementary Table 1). Prime MM-GBSA analysis disclosed the relative binding energies of these cannabinoids to 5DFP as -45.361 kcal/mol, -45.093 kcal/mol, -46.470 kcal/mol, -44.633 kcal/mol, -47.970 kcal/mol, -40.275 kcal/mol and -48.539 kcal/mol, correspondingly (Supplementary Table 1).
Nikfarjam, et al., [45] demonstrated that CBD and THC inhibited pancreatic cancer progression moderately through inhibition of PAK1. Considering this preliminary in silico study of different cannabinoids we suggest that compounds H4 CBD-3C (3d), CBD-3C (8b), HHC-1C (21f), HHC-3C (23a), HHC-5C (24b), D8 THC-3C (33d) and D8 THC-5C (34c) and CBN-7C (40) could be good inhibitors of PAK1 and therefore could be used in the treatment of pancreatic cancer.
CB 1 (5U09, 6KQI, 7V3Z) and CB 2 (5ZTY, 6KPC, 6PT0): Since CB 1 and CB 2 receptors have been discovered as meaningful molecule targets for some common disorders, the identification and design of new modulators for CB 1 and CB 2 are crucial.
The in-silico study of the interactions of cannabinoids with CB 1 and CB 2 receptors occupies a prominent place in the discussion of the agonist, antagonist and positive or negative allosteric modulator activity of these ligands on the receptors.
Allosteric ligands have been studied in the last 20 years because they present better receptor selectivity and potency than orthosteric ligands due to allosteric positions are less preserved across proteins and the opposition with endogenous is eliminated [46-48]. Allosteric modulators can be Positive Allosteric Modulators (PAM) or NAM [49]. A PAM improves the affinity, potency and/or efficacy of the ligand whereas a NAM decreases the affinity, potency and/or efficacy of the ligand [50].
In this work, we selected 7DY and AM10257 as antagonist reference ligands of 5U09 (CB 1 receptor) and 5ZTY (CB 2 receptor) respectively. CP55940, E3R and WIN 55,212-2 as agonist reference ligands of 6KQI (CB 1 receptor), 6KPC (CB 2 receptor) and 6PT0 (CB 2 receptor) respectively. CP,55940 as a negative allosteric modulator of 7V3Z (CB 1 receptor).
Supplementary Table 2-8 show the interactions between amino acids on the protein mentioned above and functional groups on tested cannabinoids, display the docking scores and exhibit the prime MM–GBSA energies.
For protein 5U09, which is bound to the antagonist 7DY of CB 1 receptor, the highest docking score is -10.321 kcal/mol corresponding to CBG-5C (14) and the lowest is -4.770 kcal/mol for CBG-3C (13) as shown in Supplementary Table 1. CBG-5C (14) exhibited multiple interactions type hydrogen-bond with the residues Ser383 and Met 103 via OH groups in the aromatic ring. Also showed π - π stacking interaction between aromatic ring-A and Phe268 amino acid residue (Supplementary Figure 3). These residues are fragments of the deep binding pocket and they are crucial for effective ligand binding. These interactions are similar to those shown by 7DY, a known CB 1 receptor antagonist. In addition, the CBG-5C: 5U09 complex was found with -52.341 kcal/ mol MM–GBSA: MMGBSA dG Bind (NS) being the best relative binding energy complex (Supplementary Table 2).
For protein 5ZTY, which is bound with AM10257 an antagonist of the CB 2 receptor, the results show that twenty-five cannabinoids of forty docked cannabinoids interacted with the amino acid residues of this protein (Supplementary Table 4). The best docking score is -10.009 kcal/mol which corresponds to CBG-5C (14) and the worst is -4.770 kcal/mol for CBG-3C (13) (Supplementary Table 4). However, CBG-5C (14) only displayed an H-bond interaction with Leu182, which is not a key residue in the binding pocket of the CB 2 receptor. The cannabinoids: 5ZTY complexes that presented the stronger relative binding energies, good docking scores and multiple interaction types π - π stacking and H-bond with the residues are H4CBD-7C (5c), CBD-1C (6a, 6b), THCBC-5C (16a), CBC-5C (19b), HHC-1C (21a, 21d, 21g, 21h), HHC-C3 (23b, 23g), HHC-C5 (24b), D9 THC-3C (28a), D8 THC-1C (29b), CBN-1C (36) and CBN-7C (40). Phe87, Phe183 and Trp194 were the most relevant amino acids in the binding pocket. The residues implied in these cannabinoid bindings match those identified in the AM10257 antagonist-binding motif. The interactions took place in the resorcinol moiety and phenolic groups. Interestingly, HHC-1C (21g) is the only ligand that interacts via π - π stacking and H-bond as shown in the 3D diagram in Supplementary Figure 4.
These in silico results demonstrate that THCBC (16a), CBC (19b), HHCs (1C-21h, 3C-23b, 5C-24) and D9 THC-3C (28a) have the most promising interactions with 5ZTY and could be possible antagonists of the CB 2 receptor.
The results from our docking study with protein 6KQI with bound CP55940 ligand as an orthosteric agonist of CB 1 receptor established that 30 cannabinoids successfully docked into the binding pocket of this protein (Supplementary Table 5). The cannabinoids that exhibited multiple interactions with amino acid residues of 6KQI, greater binding energy for 6KQI protein and a docking score higher than -7 kcal/mol were THCBG-5C (11a), CBG-5C (14), THCBC-5C (17a), CBC-5C (19b), HHC-3C (23a), D9 THC-1C (26b), D9 THC-adamantyl (27b) D9 TH-3C (28b), D9 THC-5C (29b, 29c, 29d), D8 THC-5C (33c, 33d) and CBN-C7 (40). These cannabinoids interacted with Phe170 and Phe268 forming a π - π stacking bond and with Ser383 forming a hydrogen bond in similar patterns to CP55940. CBC-5C (19b) displayed the highest docking score with -10.003 kcal/mol (Supplementary Table 5), the strongest relative binding energy complex CBC-5C (19b):6KQI (MMGBSA dG Bind (NS)=-75.939 kcal/mol: Supplementary Table 5) and four interactions with the amino acid residues of 6KQI protein in the binding pocket as shown in the 3D diagrams of Supplementary Figure 5.
Previous mutagenesis studies have established Phe170, Phe268, Leu193 and Ser383 as essential amino acids for the binding of THC analogs or related agonists such as CP55940. These amino acids interact or are close to the preferred docking pose of the ligand [51].
The results of the docking with 6KPC protein which E3R agonist bound CB 2 receptor displayed that 29 of the 40 docked cannabinoids showed good docking affinity in the binding pocket of 6KPC protein having a docking score in the range of -6.912 kcal/mol (compound 21d) to -10.557 kcal/mol (compound 27b). The most relevant amino acids in the binding pocket that interact with the aromatic ring and phenolic groups of cannabinoids are Phe87, Phe183, Thr194 via π - π stacking bond and Ser285, Ile110, Thr114 via H-bond (Supplementary Table 6).
H4 CBD-adamantyl (2c, 2d, 2e), H4 CBD-3C (3d), CBD-adamantyl (7a), CBD-3C (8a, 8d), CBD-5C (9a), THCBG (11b), THCBG-adamantyl (12b), CBG-3C (13), CBG-5C (14), CBG-adamantyl (15), THCBC-5C (17a), HHC-1C (21f, 21g), HHC-adamantyl (22f), HHC-3C (23b, 23c, 23g), HHC-5C (24f, 24c), D9 THC-adamantyl (27b), D9 THC-3C (28a, 28b), D9 THC-5C (29b, 29d), D8 THC-1C (31a, 31c, 3d), D8 THC-3C (33b, 33c, 33d) and CBN-7C (40) exhibited multiple interactions with the residues of 6KPC protein and good relative binding energies ligand: 6KPC (in the range of -52.082 kcal/mol to -79.316 kcal/mol). The most promising cannabinoids to bind with 6KPC protein are H4 CBD-adamantyl (2d) and D9 THC-adamantyl (27b) for presenting the best docking scores (-9.331 kcal/mol, -10.557 kcal/mol: Supplementary Table 5), the strongest relative binding energies (MMGBSA dG Bind (NS): -79.316 kcal/mol, -79.147 kcal/mol, respectively: Supplementary Table 6) and interacting with Phe 183, Phe 87 and Ser 285 amino acids in the binding pocket of the 6KPC protein via π - π stacking bond and H-bond (Supplementary Figure 6) which are similar to the interactions of reference agonist E3R of CB 2 receptor.
The docking studies of 40 cannabinoids with 6PTO, a Gi signaling complex bound with an agonist WIN 55,212-2 of the CB 2 receptor revealed that 17 of the docked cannabinoids interact with Phe183 and Trp194 through hydrophobic interaction. In addition, they exhibited interactions through hydrogen bonds with Thr114, Ser285 and Ile110. These interactions are similar to those shown by the well-known WIN 55,212-2-CB 2 agonist that was used as reference. The compounds that stood out with more interacting groups and stronger included H4 CBD-adamantyl (2a, 2b, 2c), CBG-3C (13), CBG-5C (14), HHC-1C (21a, 21d, 21g, 21h), HHC-3C (23a, 23g, 23h), D9 THC-1C (26c), D9 THC-5C (29c), D8 THC-1C (31c) and D8 THC-3C (33c) (Supplementary Table 6). These cannabinoids presented a docking score ranging between -5.033 kcal/mol (CBG-C3, 13) and -9.529 kcal/mol (CBG-C5, 14) (Supplementary Table 6). D8 THC-3C-:6PTO complex presented -77.056 kcal/mol, the strongest MMGBSA dG Bind (NS) among the docked cannabinoids and interact with three residues of 6PTO protein: Trp194, Phe183 through π - π stacking bond and Ile110 via and H-bond as shown 3D diagram of Supplementary Figure 7.
Ross, et al., [52] reported 7DY as the first negative allosteric modulator of CB 1. Although this compound was not approved by the FDA as a drug, has been used as a model to distinguish the allosteric site showing an uncommon complex allosteric profile at CB 1. We carried out the docking study of cannabinoids using the protein 7V3Z as a CB 1 receptor with a negative allosteric modulator 7DY bound. The specific interactions among the docked cannabinoids and 7V3Z residues are disclosed in Supplementary Table 8. The cannabinoids:7V3Z complexes that presented good affinity in the binding pocket with relative binding energies higher than -60 kcal/mol and the highest docking score (-6.981 kcal/mol to -10.821 kcal/mol) involve H4 CBD-7C (5c), CBD-adamantyl (7a), CBD-3C (8a, 8d), CBD-5C (9a), THCBG -5C (11a), THCBC-3C (16b), THCBC-5C (17a), THCBC-adamantyl (18b), CBC-5C (19a), HHC-adamantyl (22f), HHC-7C 25f), D9 THC-5C (29b, 29d) and CBN-7C (40). In addition, the most important amino acids found in the binding pocket that interact with cannabinoids include Phe170, Phe268 via π - π stacking and Ser505 via hydrogen bond (Supplementary Figure 8). It is interesting to note that THCBC-adamantyl (18b) was the ligand with the strongest relative binding energy at -86.054 kcal/mol (Supplementary Table 7).
Considering the docking study carried out using different models of CB 1 and CB 2 receptors, we demonstrated that the aromaticity of resorcinol moiety is essential for robust hydrophobic π - π stacking with amino acid residues establishing the deep binding pocket of the CB 1 and CB 2 receptors. For the three models of CB 1 receptor, these residues are Phe170, Phe268 and Trp279, which are stationed neighboring the resorcinol ring of the tested compounds. However, Phe87, Phe183 and Trp194 of the CB 2 receptor bend to and make stable the ligand binding through π - π stacking interactions with the phenolic ring-A of cannabinoids. In both CB receptors, the hydrophobic interactions principally contribute to the good docking affinity.
The aromatic hydroxyl groups at the resorcinol ring have an essential function for the CB 1 and CB 2 receptor activity. Huffman, et al., [53] reported that the substitute of the phenolic hydroxyl group in THC derivatives drastically reduces the CB 1 activity. Our docking experiments exposed the role of the hydroxyl groups in the interactions with the amino acids in the binding pocket. For CB 1 most of the cannabinoids presented hydrogen bonds between OH groups in ring A with Ser383 or Ser505, which are key interacting residues for the CB 1 affinity [54,55]. For CB 2, the cannabinoids that were docked presented phenolic group interactions with Ser285, Ile110 and/or Thr114 via hydrogen bonds. These bindings may stabilize the π - π stacking interaction with Trp194 [56].
GPR119 (7WCM): MBX-2982 is bound to 7WCM as an agonist of GPR119. Agonists that selectively activate GPR119 can be used for the treatment of metabolic disorders [57,58]. In this work, we docked 40 cannabinoids into 7WCM protein to investigate the effectiveness of the binding of cannabinoids with GPR119. Supplementary Table 9 display that CBD-1C (6a), CBG-3C (13), CBG-5C (14), THCBC-5C (17a), HHC-1C (21f, 21h), HHC-3C (23b), HHC-5C (24f), HHC-7C (25f) have multiple interactions with the amino acids of 7WCM protein, the docking scores for these cannabinoids are highest than -7.233 kcal/mol (Supplementary Table 9) and the relative binding energies of the complexes ranging between -44.477 kcal/mol (HHC-1C (21f): 7WCM) and -68.485 kcal/mol (THCBC-5C (17a): 7WCM) as displayed Supplementary Table 9. The most typical interactions are π - π stacking with Trp265 and Phe241 and hydrogen bonds with Val85 and Gluc261. Supplementary Figure 9 exhibits 3D (A,B) ligand interaction diagram of THCBC-5C (17a) and HHC-7C (25f). Considering this study, THCBC and HHC analogs presented strong relative binding energies as well as multiple interactions in the binding pocket and hence may be possible candidates to treat diabetes.
In silico ADME properties of cannabinoids
Since lack of efficacy and safety are some of the most frequent causes of why a compound does not become an approved drug, the Absorption, Distribution, Metabolism and Excretion (ADME) properties should be evaluated in the early stage of drug development. The drug-likeness and physiochemical properties of cannabinoids with docking affinity were analyzed via Maestro’s QikProp Schrodinger software. The predicted ADMET properties and descriptors for the compounds are presented in Table 1. Some cannabinoids have solubility values out of the recommended range (compounds 2,5,7,11,12,18,19,20,22,24,25,27,29,34,36,40). The solubility of cannabinoids is a challenge due to their lipophilic character. Cannabinoids with longer alkyl chains displayed poor solubility. Most other descriptors are within the recommended range by QikProp for 95% of known oral drugs. These results suggest that some of the tested cannabinoids exhibited acceptable physiochemical properties.
| Compound | MW | QPlogSa | QPlogHERGb | QPPCacoc | QPlogBBd | % Human Oral Absorptione |
|---|---|---|---|---|---|---|
| 1 | 262.39 | -4.51 | -3.719 | 2524.087 | -0.1515 | 100 |
| 2 | 382.59 | -7.214 | -4.193 | 2488.638 | -0.209 | 100 |
| 3 | 290.45 | -5.345 | -4.142 | 2524.743 | -0.309 | 100 |
| 4 | 318.5 | -5.976 | -4.431 | 2502.395 | -0.461 | 100 |
| 5 | 346.55 | -7.095 | -4.992 | 2471.448 | -0.6399 | 100 |
| 6 | 258.36 | -4.342 | -3.84 | 2754.611 | -0.114 | 100 |
| 7 | 378.55 | -7.024 | -4.347 | 2716.036 | -0.173 | 100 |
| 8 | 286.41 | -5.221 | -4.374 | 2710.076 | -0.283 | 100 |
| 9 | 314.47 | -5.583 | -4.293 | 2748.779 | -0.396 | 100 |
| 11 | 320.51 | -6.699 | -5.415 | 1747.548 | -1.093 | 100 |
| 12 | 384.6 | -7.788 | -5.124 | 1785.852 | -0.813 | 100 |
| 13 | 288.43 | -5.614 | -4.934 | 2111.985 | -0.687 | 100 |
| 14 | 316.48 | -6.463 | -4.371 | 2370.134 | -0.802 | 100 |
| 15 | 380.57 | -5.441 | -3.279 | 2808.661 | -0.327 | 100 |
| 16 | 290.45 | -6.284 | -4.871 | 3678.696 | -0.315 | 100 |
| 17 | 318.5 | -7.125 | -5.189 | 3677.395 | -0.46 | 100 |
| 18 | 382.59 | -8.041 | -4.767 | 3679.603 | -0.196 | 100 |
| 19 | 314.47 | -6.139 | -4.784 | 3570.273 | -0.369 | 100 |
| 20 | 378.55 | -7.656 | -4.775 | 3575.789 | -0.129 | 100 |
| 21 | 260.38 | -4.979 | -3.85 | 4521.113 | 0.193 | 100 |
| 22 | 380.57 | -7.674 | -4.258 | 4522.153 | 0.166 | 100 |
| 23 | 288.43 | -5.827 | -4.269 | 4520.384 | 0.051 | 100 |
| 24 | 316.48 | -6.709 | -4.705 | 4524.042 | -0.092 | 100 |
| 25 | 344.54 | -7.586 | -5.079 | 4511.257 | -0.235 | 100 |
| 26 | 258.36 | -5.047 | -4.084 | 4353.974 | 0.172 | 100 |
| 27 | 378.55 | -7.728 | -4.436 | 4354.904 | 0.145 | 100 |
| 28 | 286.41 | -5.889 | -4.473 | 4352.417 | 0.029 | 100 |
| 29 | 314.47 | -6.708 | -4.828 | 4350.853 | -0.112 | 100 |
| 31 | 258.36 | -4.927 | -4.014 | 4710.555 | 0.208 | 100 |
| 33 | 286.41 | -5.761 | -4.406 | 4715.432 | 0.068 | 100 |
| 34 | 314.47 | -6.621 | -4.821 | 4719.169 | -0.073 | 100 |
| 36 | 254.33 | -4.863 | -4.441 | 4288.338 | 0.164 | 100 |
| 40 | 338.49 | -7.542 | -5.662 | 4279.292 | -0.27 | 100 |
Note: Range of 95% drugs. (a): Predicted aqueous solubility [-6.5 to +0.5]; (b): Human Ether-a-go-go Related Gene (HERG) Potassium+ (K+) channel blockage (log IC50) [concern below: –5]; (c): Apparent Colorectal Adenocarcinoma-2 (Caco-2) cell permeability in nm/s [<25 poor; >500 excellent]; (d): Predicted log of the blood/brain partition coefficient [-3.0 to +1.2]; (e): Human oral absorption in Gastrointestinal (GI) [<25% is poor].
Table 1: Predicted Adsorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties.
In silico identification of metabolic sites of cannabinoids using cytochrome P450
Herein, we report in silico study of cytochrome P450 (CYP-enzymes)-mediated metabolic of 40 cannabinoids that were docked previously. CYPs are one the most critical enzymes in drug metabolism and therefore of importance in clinical pharmacokinetics.
In the drug discovery process, an early estimate of potential metabolites allows time and resources to be reduced by removing drug candidates that present toxic metabolites.
Using Schrodinger software, we determined the possible sites of interactions between cannabinoids and P-450 to estimate the most likely metabolites, therefore supporting the comprehension of the structural changes needed to achieve ideal metabolic stability. The results are shown in Supplementary Figure 10.
Considering the oxidative metabolism of natural cannabinoids by cytochrome P450, we only found data for HHC, D9-THC, CBD, CBC, CBG and CBN [59]. Watanabe, et al., [60] and later Anderson, et al., [61] and Roy, et al., [62] determined the major oxidized metabolites of these cannabinoids (Figure 5). Hydroxylation, epoxidation and quinone formation were the most typical reactions catalyzed by the P450 enzyme. The identified metabolites coincide with the oxidation active sites that were determined in the in-silico study. The hydroxylation of tricyclic cannabinoids (HHC, THC and CBN) is carried out on the C-11 and C8. Also, it occurs at the first carbon of the lipophilic chain except for CBN. The hydroxylation of bicyclic cannabinoids (H4 CBD and CBD) was accomplished at C6 on the terpene moiety, C10 on the propenyl group and C1 of the aliphatic chain of resorcinol ring. Finally, in the CBG and CBC analogs hydroxylation occurs in some CH2 carbons at the allylic chain of the molecule and the epoxidation takes place at the double bond of the allylic chain. In the case of CBG, Roy, et al., [62] demonstrated that after the epoxidation, undergo intramolecular cyclization to obtain the tetrahydrofuran ring attached to the resorcinol core 62 (Figure 5) [2,3]. The Quinone formation is achieved in the resorcinol ring.
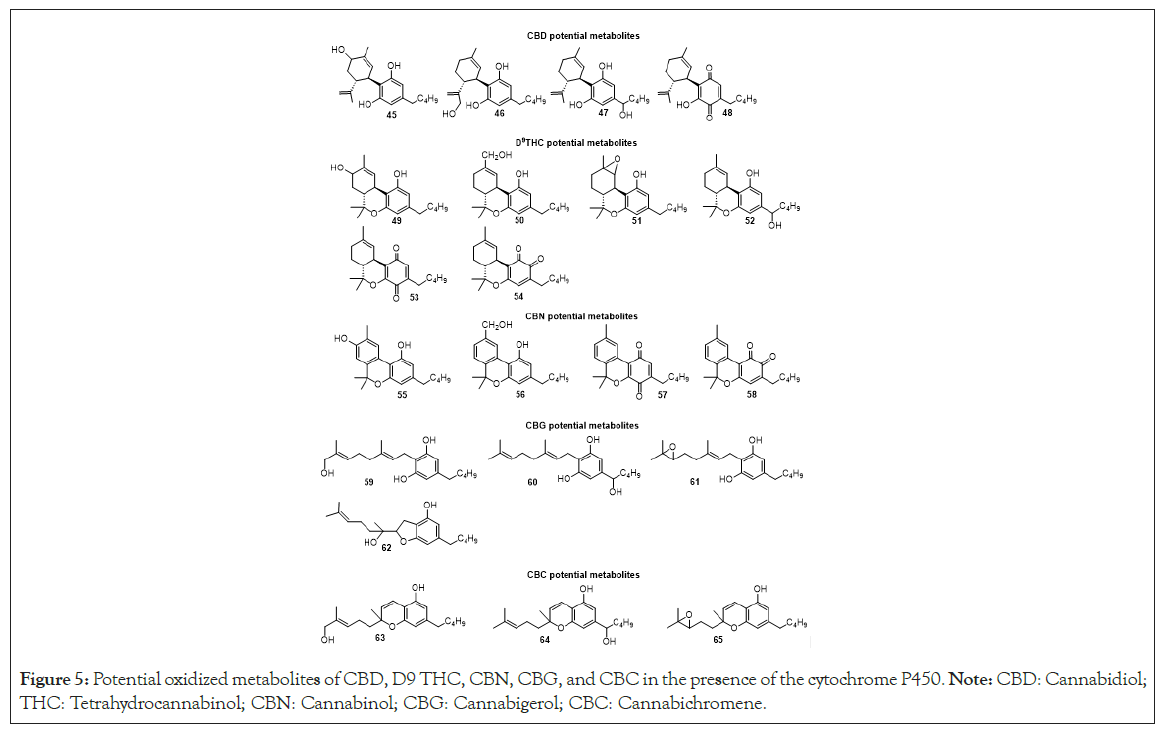
Figure 5: Potential oxidized metabolites of CBD, D9 THC, CBN, CBG, and CBC in the presence of the cytochrome P450. Note: CBD: Cannabidiol; THC: Tetrahydrocannabinol; CBN: Cannabinol; CBG: Cannabigerol; CBC: Cannabichromene.
The virtual screening residue-ligand interaction studies of saturated and unsaturated cannabinoids using different types of CB 1 and CB 2 receptors PPAR-γ and GPR119 models showed the relevance of some amino acids in the binding pocket as well as the importance of the hydrogen bond and hydrophobic interactions among cannabinoids and the residues.
The most promising cannabinoids considering docking scores, relative binding energies and multiple interactions with the protein in the binding pocket are D9 THC-1C (26b) with 2P4Y, CBN-7C (40) with 5DFP, CBG-5C (14) with 5UO9, HHC-1C (21g) with 5ZTY, CBC-5C (19b) with 6KQI, H4CBD-adamantyl (2d) and D9 THC-adamantyl (27b) with 6KPC, D8 THC-3C (33c) with 6PTO, THCBC-adamantyl (18b) with 7V3Z, THCBC-3C (17a) and HHC-7C (25f) with 7WCM. It was demonstrated that geometric constraints and lipophilicity play a crucial role in binding pockets. For example, compounds CBD-7C (10), D9 THC-7C (30) and D8 THC-7C (35) which are seven carbons in the side chain were not effectively docked into the CB 1, CB 2, GPR119, PAK1 and PPAR-γ models.
Evaluation of physiochemical properties demonstrated that the calculated properties for most compounds fall within anticipated ranges, except for cannabinoids with more than 3 carbons in the lipophilic chain, indicating suboptimal aqueous solubility. In the context of in-silico investigation into oxidative metabolism via cytochrome P450, our findings affirm that cannabinoids exhibit consistent interaction sites with CYP enzymes.
This comprehensive analysis advances our understanding of cannabinoid-protein interactions and provides valuable insights for future experimental validations and drug development endeavors.
Not applicable
There is no funding to report.
All data has been provided in the Supplemental information.
Conceptualization: Maite L. Docampo-Palacios, Giovanni A. Ramirez, Westley Cruces. Methodology: Giovanni A. Ramirez, Westley Cruces. Data Analysis: Maite L. Docampo-Palacios, Giovanni A. Ramirez, Tesfay T. Tesfatsion, Westley Cruces. Computational Modeling: Giovanni A. Ramirez. Writing – Original Draft: Maite L. Docampo-Palacios, Giovanni A. Ramirez, Westley Cruces. Writing – Revision and Editing: Maite L. Docampo-Palacios, Giovanni A. Ramirez, Tesfay T. Tesfatsion, Monica K. Pittiglio, Westley Cruces. Supervision: Westley Cruces. Project Administration: Kyle P. Ray, Westley Cruces.
All authors have read and approved this manuscript for submission.
Maite L. Docampo-Palacios, Giovanni A. Ramirez, Tesfay T. Tesfatsion and Monica K. Pittiglio are employees of Colorado Chromatography Labs. Westley Cruces and Kyle P. Ray are founders of Colorado Chromatography Labs.
Authors Maite L. Docampo-Palacios, Giovanni A. Ramirez, Tesfay T. Tesfatsion and Monica K. Pittiglio are employed by the company Colorado Chromatography Labs. Authors Kyle P. Ray and Westley Cruces are founders of the company Colorado Chromatography Labs. Authors Kyle P. Ray and Westley Cruces declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Docampo-Palacios ML, Ramirez GA, Tesfatsion TT, Pittiglio MK, Ray KP, Cruces W (2023) Decoding the Molecular Dance: In Silico Exploration of Cannabinoid Interactions with Key Protein Targets for Therapeutic Insights. J Theor Comput Sci. 9:205
Received: 28-Nov-2023, Manuscript No. JTCO-24-29219; Editor assigned: 01-Dec-2023, Pre QC No. JTCO-24-29219 (PQ); Reviewed: 15-Dec-2023, QC No. JTCO-24-29219; Revised: 22-Dec-2023, Manuscript No. JTCO-24-29219 (R); Published: 29-Dec-2023 , DOI: 10.35248/2376-130X.23.9.205
Copyright: © 2023 Docampo-Palacios ML, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Sources of funding : There is no funding to report.