
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2023)Volume 6, Issue 4
Background: Microsatellite Instability (MSI) is a clinically significant subtype in colorectal cancer. Despite the promising performance of deep learning techniques in digital pathology for clinical diagnosis, the impact of clinicopathologic factors on the performance of these models has been largely overlooked.
Methodology: Using a total of 931 colorectal cancer Whole Slide Images (WSIs), we developed and verified a deep learning algorithm and analyzed the WSI-level MSI probability and clinicopathologic variables.
Results: In both internal and external cohorts, our deep learning model achieved an Area Under the Receiver Operating Curve (AUROC) of 0.901 and 0.908, respectively. The presence of a mucinous or a signet ring cell carcinoma component enhanced the model’s ability to predict MSI (HR=19.73, P=0.026). Conversely, tumors subjected to neoadjuvant chemoradiation therapy (HR=0.03, P=0.002) and those with metastasis (HR=0.01, P=0.016) demonstrated an increased probability of being associated with Microsatellite Stability (MSS).
Conclusion: To ensure the clinical applicability of the model, it is imperative to meticulously validate deep learning- based approaches for MSI prediction, accounting for diverse practical clinicopathologic backgrounds that may impact the model’s performance.
Microsatellite instability; Microsatellite stable; Colorectal cancer; Deep learning; Whole-slide images; Mucinous carcinoma; Signet ring cell carcinoma; Neoadjuvant chemoradiation therapy
Microsatellite Instability (MSI) is a change in microsatellite length (repetitive noncoding DNA sequences) caused by a defective DNA Mismatch Repair (MMR) during DNA replication that is considered a biologically distinct molecular subtype of various cancers [1]. MSI is observed in 12-15% of all colorectal cancers. Stage II or III MSI-unstable colorectal cancer carries diagnostic, prognostic or therapeutic implications, being associated with a poor response to adjuvant chemotherapy but exhibiting a favorable stage-adjusted prognosis. Moreover, the effectiveness of immune checkpoint inhibitors for patients with advanced MSI colorectal cancer has been noted and recent prospective clinical trials have demonstrated favorable response rates of 28- 52% to PD-1 inhibitor therapies [2-4]. Thus, testing for MSI is recommended for all patients with colorectal cancer [5,6]. A pentaplex Polymerase Chain Reaction (PCR) assay has been used as the gold standard for MSI testing. Additionally, an Immunohistochemistry (IHC) panel targeting MMR proteins serves as a complementary diagnostic test to identify individual MMR gene mutations. Recently, there has been widespread adoption of MSI testing through Next Generation Sequencing (NGS). These microsatellite assays are presenting remarkable sensitivities of up to 100% for both PCR and NGS and up to 94% for Mismatch Repair Protein Immunohistochemistry (MMR IHC) [1]. However, these methods still have limitations in terms of time, cost, tissue consumption and can often be laborious.
Recently, a deep learning technique for digital images has been introduced and has shown good performance in image classification. Furthermore, research on deep learning-based digital pathologic image classification has progressed to a level where it can be applied for clinical diagnosis [7,8]. Multiple studies have developed deep learning-based classifiers for MSI status in colorectal cancer using Hematoxylin and Eosin (H&E)- stained WSIs. These classifiers have demonstrated favorable performance in a large validation cohort, surpassing the previous accuracy achieved through histology-based assessments by pathologists [7-10]. Computer-aided image analysis models can save time and money when used in actual assays, thereby reducing human labor. However, careful consideration is necessary in the clinical application of deep learning models because various clinical conditions can affect the histological features and the model’s performances. Therefore, the deep learning models used in clinical practice should be validated carefully under diverse practical clinic-pathologic backgrounds that may influence their performance [11].
In this study, we devised a deep learning framework for MSI prediction in colorectal cancer from H&E-stained images, incorporating automated tumor detection and MSI patch-level prediction (Figure 1). Using a detailed subgroup analysis, we aimed to investigate the relationship between various clinic- pathologic factors and the model’s performance.
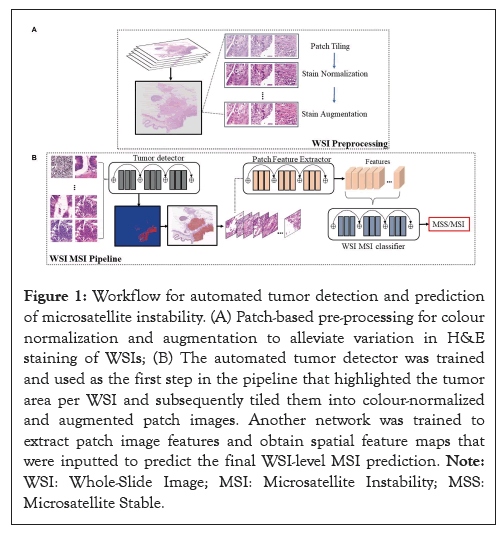
Figure 1: Workflow for automated tumor detection and prediction of microsatellite instability. (A) Patch-based pre-processing for colour normalization and augmentation to alleviate variation in H&E staining of WSIs; (B) The automated tumor detector was trained and used as the first step in the pipeline that highlighted the tumor area per WSI and subsequently tiled them into colour-normalized and augmented patch images. Another network was trained to extract patch image features and obtain spatial feature maps that were inputted to predict the final WSI-level MSI prediction. Note: WSI: Whole-Slide Image; MSI: Microsatellite Instability; MSS: Microsatellite Stable.
Patient cohorts and dataset partition
In our intra-cohort experiments, we employed a two-step cohort collection process, approved by the Institutional Review Board (IRB) of Asan Medical Center under waiver of patient informed consent (Approval No. 2019-1192) (Figure 2). Initially, we retrospectively obtained 639 resected colorectal cancer cases from Asan Medical Center (2009-2016). Pentaplex PCR was performed, excluding cases with insufficient material or quality. These cases were randomly divided into training (n=351; 231 with MSS, 120 with MSI), validation (n=90; 59 with MSS, 31 with MSI) and internal test sets (n=198; 119 with MSS, 79 with MSI).
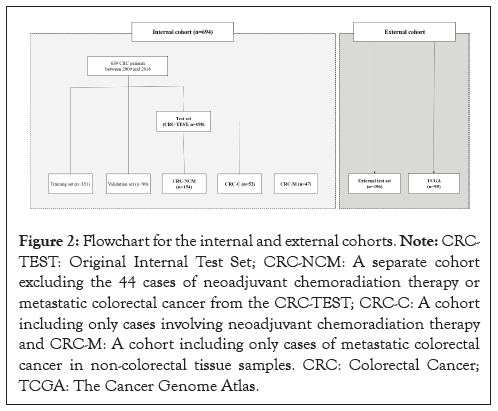
Figure 2: Flowchart for the internal and external cohorts. Note: CRC- TEST: Original Internal Test Set; CRC-NCM: A separate cohort excluding the 44 cases of neoadjuvant chemoradiation therapy or metastatic colorectal cancer from the CRC-TEST; CRC-C: A cohort including only cases involving neoadjuvant chemoradiation therapy and CRC-M: A cohort including only cases of metastatic colorectal cancer in non-colorectal tissue samples. CRC: Colorectal Cancer; TCGA: The Cancer Genome Atlas.
The Colorectal Cancer (CRC)-Test set (n=198) included 26 rectal cancer cases with neoadjuvant chemoradiation therapy and 18 metastatic colorectal cancer cases. No such cases were included in the training and validation sets. Our evaluation of the model’s performance and clinicopathologic variables using the CRC-TEST set revealed significant impacts from neoadjuvant chemoradiation therapy and metastatic colorectal cancer. To validate these findings, we performed detailed subgroup analyses with additional cases based on neoadjuvant therapy or metastasis.
To further validate, we created the CRC-NCM set (n=154, excluding 44 cases with neoadjuvant therapy or metastasis from the CRC-TEST set). Additionally, we collected 55 cases with neoadjuvant chemoradiation therapy (CRC-C, n=52) and metastatic colorectal cancer in non-colorectal tissue (CRC-M, n=47), ensuring no overlap between these sets.
For external validation, we utilized two external cohorts: an external test set from Gangnam Severance Hospital (n=196) and The Cancer Genome Atlas (TCGA) dataset (n=95). Both cohorts had digital H&E slide images from Formalin-Fixed Paraffin- Embedded (FFPE) tissue of primary resected colorectal cancer specimens, with a practical MSI to MSS ratio of 1:9. The external test set had no cases involving neoadjuvant chemoradiation therapy or metastatic colorectal cancer.
All samples in the internal and external cohorts were fully anonymized, with MSI-Polymerase Chain Reaction (PCR) serving as the ground truth following standard protocols. The distribution of microsatellite status, based on MSI-PCR results in each dataset is outlined. For additional analysis, we utilized alternative ground truth labels through MMR-IHC or targeted NGS with detailed methods.
Preparation and model development
One representative FFPE and H&E-stained tumor slide from each case was selected by manual review that was blinded to clinical information and microsatellite status. The slides were scanned at 20x magnification (0.5 μm/pixel) with an Aperio AT2 scanner (Leica Biosystems, Wetzlar, Germany) and then scanned at 20x magnification (0.242 μm/pixel) with a PANNORAMIC® 250 Flash I scanner (3D Histech Ltd., Budapest, Hungary).
Figure 1 shows the main steps for our model development: (i) WSI preprocessing followed by training an automated tumor detection model. (ii) Employment of the trained tumor detector to highlight relevant regions per WSI, extraction of the patches, and training of a secondary model for MSS/MSI patch probability prediction. (iii) WSI-level training of the model to predict MSS/MSI using patch image features extracted by the trained model (step-ii) and integration of information of all patch-based features per WSI.
Clinicopathologic characteristics of microsatellite instability
Demographic, clinical and pathologic data from 198 patients in the CRC-TEST set and 196 patients in the external test set were retrieved by reviewing medical records including age, sex, stage based on the 8th edition of the American Joint Committee on Cancer (AJCC) system, therapy with neoadjuvant chemoradiation and tumor location and size. Two experienced pathologists reviewed all WSIs from the test sets (198 WSIs from CRC-TEST and 196 WSIs from the external test set) and manually estimated the mucinous component, TILs and the presence of neutrophilic and eosinophilic infiltration, which are associated with MSI prediction [12-14]. The experts were blinded regarding MSI status and any other clinical information during review of the WSIs and further details for the criteria we used for the histologic assessments.
Statistical analysis
The model performance for MSI prediction was measured using weighted accuracy, which was calculated by considering the different fractions of MSI in each data set, sensitivity, specificity, AUROC and Area Under the Precision Recall Curve (AUPRC). The Chi-square test, Fisher’s exact test and univariate and multivariate logistic regression analyses were performed to evaluate the relationship between the predicted microsatellite status and clinicopathological characteristics. All statistical analyses were performed using SPSS software package version 21.0.0 (SPSS Statistics software, IBM Corp, NY, USA) and R version 4.0.0 (R Foundation for Statistical Computing, Vienna, Austria). A P-value < 0.05 was considered statistically significant.
Performance for tumor segmentation and microsatellite instability prediction
Performance of MSI prediction was evaluated in the subcategorized datasets based on neoadjuvant chemoradiation therapy or metastasis (Figure 2). The tumor area was automatically detected with a mean Dice value of 0.696 and a mean Intersection over Union (IoU) value of 0.741 for the CRC-TEST set and a mean Dice value of 0.674 and a mean IoU value of 0.732 for the external test set. Subsequently, our model predicted microsatellite status in the detected tumor areas of the WSIs for each case and the representative heatmaps for MSI prediction are shown in Figure 3A. Utilizing the CRC-TEST set, our deep learning model predicted MSI with an AUROC of 0.778 (95% Confidence Interval (CI): 0.705-0.851), an AUPRC of 0.783 (95% CI: 0.678- 0.860), a weighted accuracy of 0.741, a sensitivity of 0.61 and a specificity of 0.87 at the WSI level (Figure 3B and Table 1). Using the external test set, our model achieved an AUROC of 0.908 (95% CI: 0.856-0.959), an AUPRC of 0.523 (95% CI: 0.313- 0.725), with a higher weighted accuracy than that achieved on the internal test set at 0.83, a sensitivity of 0.90 and a specificity of 0.80. When applying the TCGA dataset, we observed a relatively decreased performance with an AUROC of 0.775 (95% CI: 0.680-0.871), an AUPRC of 0.189 (95% CI: 0.046-0.531), a sensitivity of 0.85 and a specificity of 0.30 (Figure 3C).
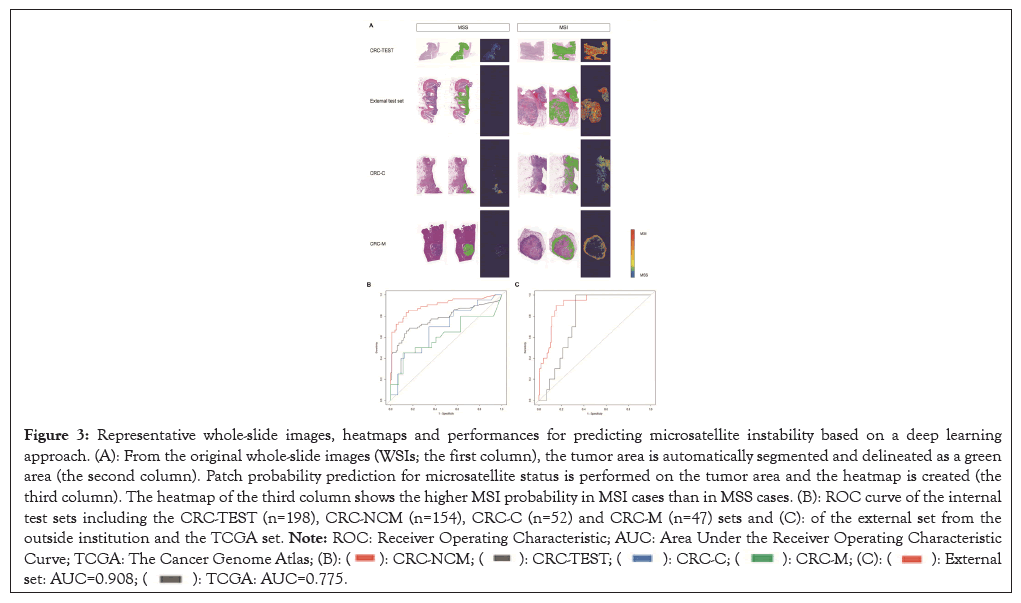
Figure 3: Representative whole-slide images, heatmaps and performances for predicting microsatellite instability based on a deep learning
approach. (A): From the original whole-slide images (WSIs; the first column), the tumor area is automatically segmented and delineated as a green
area (the second column). Patch probability prediction for microsatellite status is performed on the tumor area and the heatmap is created (the
third column). The heatmap of the third column shows the higher MSI probability in MSI cases than in MSS cases. (B): ROC curve of the internal
test sets including the CRC-TEST (n=198), CRC-NCM (n=154), CRC-C (n=52) and CRC-M (n=47) sets and (C): of the external set from the
outside institution and the TCGA set. Note: ROC: Receiver Operating Characteristic; AUC: Area Under the Receiver Operating Characteristic
Curve; TCGA: The Cancer Genome Atlas; 
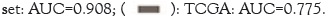
| Accuracy (weighted) | Sensitivity | Specificity | AUC | |
|---|---|---|---|---|
| CRC-TEST (n=198) | 0.741 | 0.61 | 0.87 | 0.778 |
| CRC-NCM (n=154) | 0.829 | 0.78 | 0.88 | 0.901 |
| CRC-C (n=52) | 0.692 | 0.35 | 0.91 | 0.681 |
| CRC-M (n=47) | 0.638 | 0.3 | 0.89 | 0.61 |
| External test set (n=196) | 0.83 | 0.9 | 0.8 | 0.908 |
| TCGA (n=95) | 0.789 | 0.85 | 0.3 | 0.775 |
Note: AUC: Area Under the Receiver Operating Characteristic Curve; CRC-TEST: Original internal test set; CRC-NCM: a separate cohort excluding the 44 cases of neoadjuvant chemoradiation therapy or metastatic colorectal cancer from the CRC-TEST; CRC-C: a cohort including only cases involving neoadjuvant chemoradiation therapy; CRC-M: a cohort including only cases of metastatic colorectal cancer in non-colorectal tissue samples; CRC: colorectal cancer; TCGA: The Cancer Genome Atlas.
Table 1: Performance of the deep learning model for microsatellite instability prediction on the test sets.
Clinicopathologic implications of predicted microsatellite instability
On the CRC-TEST set, the baseline clinicopathologic characteristics with microsatellite status determined using the pentaplex PCR method, which was used as ground truth, are presented in (Table 2). Variables, including age younger than 50 years, right-sided location, tumor size ≥ 6 cm, poorly differentiated tumor, mucinous or signet ring cell carcinoma, medullary carcinoma, expansile tumor growth, presence of Tumor Infiltrating Lymphocyte (intra-epithelial) (eTILs), Crohn’s- like lymphocytic reaction and the absence of dirty necrosis, were associated with an unstable MSI status.
| MSI-PCR | ||||
|---|---|---|---|---|
| Variable | MSS (n=119) | MSI (n=79) | P | |
| Age | <50 years | 24 (20.2) | 31 (39.2) | 0.003 |
| ≥ 50 years | 95 (79.8) | 48 (60.8) | ||
| Sex | Male | 92 (68.1) | 36 (57.1) | 0.131 |
| Female | 43 (31.9) | 27(42.9) | ||
| Anatomic site | Right-sided | 23 (20.0) | 33 (61.1) | <0.001 |
| Left-sided | 87 (75.7) | 17 (31.5) | ||
| Transverse | 5 (4.3) | 4 (7.4) | ||
| Tumor size | ≥ 6 cm | 39(32.8) | 27 (50.0) | 0.031 |
| <6 cm | 80 (67.2) | 27 (50.0) | ||
| T stage | Tis/T1 | 2 (1.8) | 5 (7.4) | 0.089 |
| T2 | 6 (5.4) | 8 (11.8) | ||
| T3 | 74 (66.7) | 37 (54.4) | ||
| T4a-b | 29 (26.1) | 18 (26.5) | ||
| Histology | Adenocarcinoma, WD | 7 (5.9) | 0 (.0) | <0.001 |
| Adenocarcinoma, MD | 95 (79.8) | 42 (53.2) | ||
| Adenocarcinoma, PD | 11 (9.2) | 15 (19.0) | ||
| Mucinous or signet ring cell carcinoma | 3 (2.5) | 14 (17.8) | ||
| Medullary carcinoma | 3 (2.5) | 8 (10.1) | ||
| Expansile growth | Present | 8 (7.5) | 27 (37.0) | <0.001 |
| Absent | 99 (92.5) | 46 (63.0) | ||
| eTILs | Present | 37 (31.1) | 61 (77.2) | <0.001 |
| Absent | 82 (68.9) | 18 (22.8) | ||
| Crohn’s-like lymphocytic reaction |
Present | 60 (52.2) | 51 (67.1) | 0.041 |
| Absent | 55 (47.8) | 25 (32.9) | ||
| Necrosis | Lack of dirty necrosis | 14 (11.8) | 47 (59.5) | <0.001 |
| Presence of dirty necrosis | 105 (88.2) | 32 (40.5) | ||
| MMR IHC | Loss of expression in one or more MMR proteins | 3 (2.5) | 46 (85.2) | <0.001 |
| No loss | 116 (97.5) | 8 (14.8) | ||
Note: MSS: Microsatellite Stable; MSI: Microsatellite Instability; PCR: Polymerase Chain Reaction; WD: Well-Differentiated; MD: Moderately Differentiated; PD: Poorly Differentiated; eTILs: Tumor-Infiltrating Lymphocytes (intra-epithelial); MMR IHC: Mismatch Repair Protein Immunohistochemistry
Table 2: Clinicopathologic features and microsatellite status of polymerase chain reaction on the CRC-TEST set.
Using univariate logistic regression analysis, we further demonstrated that variables, including right-sided location, size ≥ 6 cm, poor differentiation, mucinous or signet ring cell carcinoma, medullary carcinoma, expansile growth, presence of eTILs and the absence of dirty necrosis, were significantly associated with the model’s MSI prediction performance on the CRC-TEST set. These results suggest that histopathological changes typically associated with MSI also influence the MSI prediction performance of the model. Through an analysis that categorized cases into three groups based on the content of a mucinous or signet ring cell carcinoma component, the group with a higher mucin component (≥ 50%) showed significantly higher Hazard Ratio (HR) than those of the groups with lower mucin components in both the internal and external sets (CRC-TEST, HR=18.60, P=0.029 and the external test set, HR=5.11, P=0.048).
Multivariate logistic regression analysis on the CRC-TEST set revealed that only the variables of mucinous or signet ring cell carcinoma (HR=19.73, P=0.026), neoadjuvant chemoradiation therapy (HR=0.03, P=0.002) and metastasis (HR=0.01, P=0.016) were significant independent factors for predicting MSI status (Table 3). In the external set, variables including poor differentiation, medullary carcinoma, Crohn’s-like lymphocytic reaction, neoadjuvant chemoradiation therapy and metastatic colorectal cancer were excluded from the multivariate analysis due to an insufficient number of positive samples. The presence of a mucinous or signet ring cell carcinoma component reinforced the model’s ability to predict MSI, while neoadjuvant chemoradiation therapy and metastasis reinforced the model’s ability to predict MSS.
| CTC-TEST set | External test set | |||
|---|---|---|---|---|
| Covariates | HR (95% CI) | P value | HR (95% CI) | P value |
| Poorly differentiation | 0.88 (0.219-3.52) | 0.854 | - | - |
| Medullary carcinoma | 8.79 (0.69-112.12) | 0.094 | - | - |
| Expansile growth | 3.27 (0.71-15.16) | 0.129 | 2.45 (0.76–7.97) | 0.135 |
| eTILs | 1.29 (0.39-4.25) | 0.679 | 2.24 (0.94–5.33) | 0.068 |
| Crohn’s-like lymphocytic reaction | 0.37 (0.12-1.14) | 0.085 | - | - |
| Necrosis | 0.95 (0.30-2.99) | 0.932 | 0.36 (0.15-0.85) | 0.02 |
| Mucinous or signet ring cell carcinoma | 19.73 (1.44-270.30) | 0.026 | 1.03 (1.01-1.05) | 0.004 |
| Neoadjuvant chemoradiation therapy | 0.03 (0.00-0.26) | 0.002 | - | - |
| Metastatic colorectal cancer | 0.01 (0.00-0.45) | 0.016 | - | - |
| MSI-PCR | 21.51 (6.21-74.54) | <0.001 | 24.97 (4.82-129.48) | <0.001 |
Note: eTILs: Tumor-Infiltrating Lymphocytes (intra-epithelial); MSI: Microsatellite Instability; PCR: Polymerase Chain Reaction; HR: Hazard Ratio; CI: Confidential Interval.
Table 3: Multivariate logistic regression analysis for microsatellite instability prediction.
Additionally, we investigate the distribution between the predicted MSI and MSS groups using manual assessment of Stromal Tumor- Infiltrating Lymphocytes (sTILs), neutrophilic infiltration and eosinophilic infiltration on WSIs. Only a neutrophilic infiltration (>50/HPF) was significantly associated with MSI prediction rather than MSS in the CRC-TEST set (P=0.002); this finding was not observed in the external test set. Otherwise, sTILs and eosinophilic infiltration were not significantly associated with MSI prediction in both CRC-TEST and external test sets. Reviewing the patch images of WSIs that were classified as MSS or MSI, we found common histologic features within each group of microsatellite status, despite varying degrees of histo-morphologic heterogeneity. Stromal inflammatory cell infiltration, encompassing mononuclear cells, neutrophils and eosinophils, as well as the presence of a poorly differentiated area and mucinous components, were more pronounced in cases predicted to be MSI than in those predicted to be MSS.
Performance of predicting microsatellite instability on tumors with neoadjuvant therapy or metastasis
To confirm the effect of neoadjuvant chemoradiation therapy or metastasis on the model’s performance for predicting MSI, additional test sets (CRC-NCM, n=154; CRC-C, n=52; CRC-M, n=47) were constructed. In the CRC-C or CRC-M sets, the model tended to classify each patch as MSS instead of MSI (Figure 3A). This was also confirmed by the multivariable analysis as described above (Table 3). On the CRC-NCM set, the model achieved a significantly higher performance than on the CRC-TEST set with an AUROC of 0.901 (95% CI: 0.845-0.958), an AUPRC of 0.877 (95% CI: 0.759-0.941), an accuracy of 0.829 and a sensitivity of 0.78 (Figure 3B and Table 1). Of note, the performance gap in the AUROC between the CRC-NCM and CRC-TEST sets was statistically significant (P=0.010). We observed a decline in performance on the CRC-C and CRC-M sets with AUROC values of 0.681 (95% CI: 0.530-0.832) and 0.610 (95% CI: 0.436- 0.785), respectively. The corresponding AUPRC values were 0.565 (95% CI: 0.349-0.758) for the CRC-C and 0.613 (95% CI: 0.392-0.796) for the CRC-M. The model exhibited a notable decrease in sensitivity, with values of 0.35 on the CRC-C set and 0.3 on the CRC-M set.
Incorporating the deep learning-based assay with mismatch repair protein immunohistochemistry
We further explored the model’s performance by combining our deep learning-based model with MMR IHC, presenting a method of utilizing the model as a diagnostic assistant or screening tool. The combined tools for MSI measurement were tested on the CRC-NCM set with the ground truth of MSI-PCR. Beginning with the model’s prediction, cases with predicted MSS and retained MMR expression are classified as the MSS group and cases with predicted MSI or loss of MMR expression are classified as the MSI group. This incorporated prediction model achieved improved performance, with a sensitivity of 0.94 and a similar specificity of 0.86, compared with the performance of the model’s prediction alone.
Performance on the modified ground truth using targeted next-generation sequencing
Reviewing the discordant results between MSI-PCR and the model’s prediction, we identified 9 cases that exhibited a substantial disparity in microsatellite status between MSI-PCR and the probability scores calculated by the model: MSS on MSI- PCR but probability scores of >0.98 on the model’s prediction (4 cases) or MSI on MSI-PCR but probability scores of approximately zero (5 cases). Subsequently, we conducted targeted NGS on these 9 cases and discovered that among the cases initially identified as MSI-positive by MSI-PCR, 3 cases were later found to be false- positive results due to interpretation errors. Upon reevaluation, these cases were re-classified as MSS. With the modified ground truth determined by NGS, the model’s performance yielded an increased AUROC of 0.910 (95% CI: 0.853-0.967), an increased sensitivity of 0.80 and a similar specificity of 0.87 compared with those obtained with the original ground truth of MSI-PCR.
The prediction of MSI or mismatch repair deficiency in colorectal cancer using H&E stained histology slides has emerged as an intriguing and current issue in digital pathology. Multiple studies have introduced new algorithms for MSI prediction, demonstrating remarkable performances [7-9,11,15,16]. In this study, our primary objective was to develop and externally validate a model for predicting MSI, achieving an AUROC of 0.908, a performance level comparable to that reported in previous studies. Furthermore, we investigated the impact of various clinicopathologic parameters on the model’s performance and found that predictions on tumours with neoadjuvant therapeutic effects or those with metastasis significantly hinder the model’s accuracy.
While previous studies have explored the connection between clinicopathologic features and deep learning-based MSI prediction, these features were either not easily understandable or had minimal impact on the model’s outcome [7,11]. Recent studies have revealed that several morphologic features within tumours such as debris, lymphocytes, or necrotic cells are significantly associated with MSI prediction. However, it is important to note that these features may also have a possibility of false positivity when predicting MSI [10,17]. Echle, et al., [16] demonstrated that mucinous carcinoma was significantly associated with false positivity, indicating that extensive mucinous tumors should be validated using conventional standard methods of MSI testing rather than relying solely on AI systems. Even though previous studies have evaluated only a subset of histologic features, it is crucial to recognize that clinicopathologic variables encountered in real-world have a risk of misclassification and should not be disregarded.
We showed that multiple MSI-associated clinicopathologic parameters (right-sided tumour location, size ≥ 6 cm, poor differentiation, mucinous or signet ring cell carcinoma, medullary carcinoma, expansile growth, presence of eTILs, and lack of dirty necrosis) were significantly associated with predicted MSI. The presence of mucinous or signet ring cell carcinoma showed an increase in the HR in proportion to the content ratio, suggesting that the presence of mucinous component might enhance the model’s predictive ability for MSI (Table 3). It is worth noting that variables of neoadjuvant chemoradiation therapy and metastasis have shown a significant negative relationship with MSI prediction, indicating that they can act as confounding factors for the model’s performance.
In the CRC-C set (patients who underwent neoadjuvant chemoradiation therapy), there were 10 cases of false negativity (accounting for 45.5% of the predicted MSS group) without any false positivity. When reviewing the patch images, no discernible histo-morphologic differences were observed between the true negative and false negative groups in the CRC-C set. Despite the presence of histologic changes by therapeutic response, such as predominant colloid change, nuclear atypia, eosinophilic cytoplasmic change and fibrous or inflammatory stromal change, these features did not appear to contribute to distinguishing between true negatives and false negatives. Therefore, the high tendency of false negativity in the CRC-C set may be attributed to a low proportion of tumour cells detected in the slide images. A previous study showed lower than average performance for MSI prediction in patients with rectal cancer, which might be influenced by the negative impact of neoadjuvant therapy [7].
Visualizing patches in the CRC-M set (metastatic colorectal cancer), we observed the presence of several extracted patches of normal hepatic parenchyma in certain cases that were predicted as MSI. This suggests that the histo-morphologic similarity recognized by the model between the tumour cells and hepatocytes could potentially reduce the performance. These challenges might be attributable to differences in tissue types between the training/ validation, which was performed on WSIs of colorectal tissues and the CRC-M set. To overcome these limitations and optimize a deep learning model for MSI prediction in clinical practice, it is crucial to conduct a large cohort study that incorporates transfer learning beyond specific tissue types. This approach would help enhance the model's generalizability and improve its performance across diverse clinical scenarios.
Given the impact of clinicopathologic variables on the model’s outcome, relying solely on a deep learning-based approach for MSI testing may have limitations. However, it can be utilized in clinical practice alongside conventional MSI testing methods. By combining both approaches, it is possible to reduce false results and improve the overall accuracy of MSI testing. As a feasible alternative screening tool, we suggested MSI prediction-workflow diagrams where MMR IHC was added to the model’s prediction. Indeed, a recent study has suggested an approach that combines AI-based screening for MSI followed by confirmation using conventional MSI tests [9]. This strategy optimizes resources and reduces unnecessary testing, making the overall process more cost-effective while maintaining accuracy in MSI detection.
Our deep learning-based MSI prediction was based on patches within tumour areas and as a result, the extracted histo- morphologic features were primarily tumour-associated. Certain features, such as Crohn’s-like lymphocytic reaction, which is a known characteristic of MSI and mainly located outside of the tumour area, did not exhibit a correlation with our MSI prediction. In this study, scanning was performed at 20x magnification, which may have limited the incorporation of higher-resolution information such as image texture and tissue boundary details into the prediction. However, all input patches were standardized to a resolution of 0.5 μm/pixel, mitigating the impact of scanning at 20x magnification on WSIs [8]. Another limitation is the MSI proportion in the internal test set (40%), which was higher than the practical ratio of 10-20% [18]. Nevertheless, the internally trained model demonstrated better MSI prediction performance on the external test set, which consisted of a practical proportion of MSI cases (10%).
Our study explored the influence of clinicopathologic features on deep learning-based MSI prediction, an aspect that has received limited attention in prior research. We found that prediction on tumour specimens with neoadjuvant therapy or metastasis can significantly impact the model’s performance for MSI prediction. Our results highlight the necessity of meticulous validation of deep learning models under diverse clinicopathologic backgrounds to ensure their clinical applicability. Incorporating such models alongside conventional MSI testing methods can enhance accuracy and optimize resources in clinical practice. Moreover, the study underscores the importance of considering specific study components for deep learning- based MSI prediction to ensure accurate and appropriate application in real-world clinical scenarios.
This study was supported by a grant (2021IP0064) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
The authors have declared no conflicts of interest.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar ] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Kim M, Chikontwe P, Go H, Jeong JH, Shin SJ, Park SH, et al. (2023) Deep Learning for Microsatellite Instability Prediction in Colorectal Cancer: Impact of Clinicopathologic Variables on Model Performance. J Clin Chem Lab Med. 6:276.
Received: 27-Nov-2023, Manuscript No. JCCLM-23-28429; Editor assigned: 29-Nov-2023, Pre QC No. JCCLM-23-28429 (PQ); Reviewed: 13-Dec-2023, QC No. JCCLM-23-28429; Revised: 20-Dec-2023, Manuscript No. JCCLM-23-28429 (R); Published: 27-Dec-2023 , DOI: 10.35248/2736-6588.23.6.276
Copyright: © 2023 Kim M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.