Journal of Depression and Anxiety
Open Access
ISSN: 2167-1044
ISSN: 2167-1044
Research - (2022)Volume 11, Issue 10
Introduction: As the share of elderly in the population is increasing, so is the presence of depression and anxiety in this group, including in Brazil. There are studies suggesting common pathophysiological mechanisms for depressive and anxiety disorders, as well as the existence of vulnerability genes in the etiopathogenesis of both depression and anxiety. The different candidate genes reported in the literature associated with depression and/or anxiety phenotypes have rarely been investigated together in a single study.
Objective: To investigate candidate gene polymorphisms, reported as associated with a higher risk of developing depression and/or anxiety symptoms in the literature, in an elderly population.
Methodology: Peripheral venous blood was collected from a total of 874 elderly people aged 60 years or older. Genotypic DNA analysis was performed by real-time PCR of 27 polymorphisms of 11 candidate genes for symptoms of depression and/or anxiety. Depressive and/or Anxious Symptomatology Groups (DASG) was also included in the analysis based on the median of three applied scales: the CES-D for depression, the GAI for anxiety, and the MMSE for cognition. For statistical analysis, Pearson's chi-square test was performed with a significance level of 5% (p= ≤ 0.05), both for individual analysis of polymorphisms and for the joint analysis.
Results: Four polymorphisms showed statistically significant results associated with DASG: rs8071667 (p=0.03) of the 5HTT gene, rs6265 (p=0.004) of the BDNF gene, rs165599 (p=0.023) of the COMT gene, and rs1417938 (p=0.006) of the CRP gene. The rs165599 (COMT) and rs1417938 (CRP) variants remained significant when analyzed together, with a p-value of 1.72E-10. Conclusion: The COMT gene variant rs165599 and CRP gene variant rs1417938 provided the most robust results in our analysis. However, it is necessary to confirm the reproduction of these preliminary results in independent samples.
Mental disorder; Genetic variants; Vulnerability; Susceptibility
In recent decades, there has been an increase in the population aged over 60 years in relation to the general population, and thus, as in many other countries, the percentage of older people in the Brazilian population has increased, currently comprising 28 million older people [1,2]. In addition to the appearance of several pathologies in this age group, depression and anxiety are reported as two of the main mental disorders in older adults [3-5].
According to the World Health Organization (WHO), depression is a public health problem [4,6,7], affecting approximately 264 million people, including older adults, and it is also one of the main causes of disability in life, generating emotional suffering and worsening quality of life [4,6,7]. In a 2019 Brazilian survey, 13.2% of Brazilian older adults between 60 and 64 years of age presented depression–(IBGE 2018), with a greater prevalence in women [8-10].
Depressive disorders present heterogeneity in the presentation of symptoms [4,6,7], where the most common symptoms are: sleep and weight alterations, feelings of worthlessness and guilt, fatigue, loss of energy, agitation or psychomotor retardation, lack of concentration, and also suicidal ideation [11]. In addition, in older people, the symptoms of depression can often be confused with dementia, and careless clinical observation in older adults can result in inadequate treatment [12,13].
Anxiety disorder is also a widespread problem, which is already a prevalent disorder for this age group, contributing to the reduction in quality of life, and restricting social life and independence [14- 16]. The most common symptoms of anxiety disorder are: fear, anguish, excessive worry, restlessness, sleep disturbances, irritability, difficulty concentrating, muscle tension, and tiredness [17].
There are several reports on the comorbidity of depression and anxiety, that is, both disorders can be present together in up to 50% of cases [16,18]. Naturally, the combination of depressive and anxious symptoms can contribute to worsening of the general clinical framework, disabling these patients to a greater extent, with larger physical, social, and psychological damages, which can even make treatment difficult [16,19].
The participation of the genetic component is present for both depressive disorder and anxiety disorder [20]. The presence of a gene or a set of them in the pathological mental process has been investigated in several studies, including in studies of the phenotypic association of only depressive or anxious symptoms with genetic markers [20].
It is known today that the relationship of genes in the development of depressive and anxious symptoms is complex, and that these disorders are also influenced by environmental factors (cultural, psychological, among others) in addition to genetic factors [21,22]. The genetic heritability for depressive disorder is estimated to be around 37% [23], while for anxiety it is around 35% [24].
More recent association studies have scanned the entire genome of an individual, for example, a million genetic markers, although the results are still inconclusive. In addition, there are a very small number of molecular genetic studies in the older population (over 60 years of age) [24].
Based on the international literature, we initially selected 11 genes that were associated with psychiatric disorders, including depression and/or anxiety, as follows: (a) the gene encoding the serotonin transporter protein (5HTT or SLC6A4) has been selected as a candidate gene in several studies investigating various psychiatric phenotypes [25,26]. The involvement of serotonin transport has been proposed as an important factor in the development of depressive disorders [25,26]. ; (b) The gene for Apolipoprotein E (APOE), a lipoprotein transport glycoprotein that is involved with susceptibility to Alzheimer's Disease (AD) [27,28]. Studies have reported that the epsilon 4 allele, in addition to being a risk factor for the development of AD, also contributes as a risk factor for depression [29,30]; (c) the cardiovascular system was also associated with a risk for the development of depression, the gene AGTR1 (Angiotensin II Receptor 1) is the most studied for this association [31,32]. Some studies report that depression and dementia share similar risk factors and common pathogenetic history, including cardiovascular risk factors [31]; (d) another gene widely investigated in the literature is the Brain-Derived Neurotrophic Factor (BDNF), the most studied polymorphism is rs6265, known as Val66Met (where a guanine base is replaced by adenine at position 196, leading to the exchange of the amino acid valine for methionine at codon 66). There are reports in the literature associating this substitution of valine for methionine with susceptibility to the development of psychiatric disorders, including depressive disorders [33-36]; (e) the Catechol-O-Methyl Transferase (COMT) gene is another relevant research target for depression and anxiety disorders. The COMT-encoded enzyme plays a key role in the degradation of catecholamines (adrenaline, noradrenaline, and dopamine) [37,38]. Previous studies indicate that the alteration in these neurotransmissions of the dopaminergic system can lead to some symptoms of depressive frameworks [39,40]; (f) some studies have suggested that folate deficiency, a cofactor in homocysteine metabolism, can lead to hyperhomocysteinemia, and this enzymatic insufficiency would be associated with the rs1801133 (C677T) and rs1801131 (A1298C) polymorphisms of the gene MTHFR (MethyleneTetraHydroFolate Reductase), suggesting that as a result, these polymorphisms may be associated with depression [41-43]; (g) it has also been proposed that alterations in the signaling of the pathways of the target protein of rapamycin (MTOR) are involved in difficulties in the mechanisms of learning, memory and also, psychiatric disorders, including depressive symptoms [44-46]. Finally, some studies have associated increased levels of inflammatory cytokines and their soluble receptors in peripheral blood in individuals with anxiety, mood, and depressive disorders [47-49].
We investigated cytokines that were proposed by some studies as a risk factor for depression and/or anxiety, being (h) Interleukin 6 (IL-6), (i) Interleukin 10 (IL-10), (j) C-Reactive Protein (CRP), and (k) Tumor Necrosis Factor (TNF) [50-55].
After selecting the 27 genetic markers of the 11 candidate genes described above, an association study was carried out between these markers in a population-based design, involving older people aged 60 years or over, and their correlation with depressive and/ or anxious symptoms. A cognitive assessment was included in the analysis, as alterations in cognition are frequently reported in older adults (presence of cognitive deficit or dementia).
Clinical methodology
Older individuals who were attended by Basic Health Units (UBS) in the Butantã region in the city of São Paulo, Brazil were selected for the study. Initially, all the older adults (individuals aged 60 years or older) were screened for subsyndromal symptoms of depression and/or anxiety. All participants signed an informed consent form and the project was approved by the local research ethics committee of the Hospital das Clínicas-Faculdade de Medicina da Universidade de São Paulo (HC-FMUSP)–05403-000, FAPESP number-2012/50010-0.
For the screening of depressive symptoms, we used the CES-D (Center for Epidemiologic Studies Depression Scale) [56]. This scale evaluates the feelings and behaviors of the older adult arising over the previous two weeks [57,58]. For this scale, the majority of studies use a value equal to or greater than 16 as a cut-off point, however, this is not a rule since other studies used lower cut-off points, such as 13 [59] or a higher cut-off point of 20 [60]. This decision depends on the socio-economic, cognitive, and cultural characteristics of the sample evaluated, consisting of 20 items in which the older person marks the answer declaring to agree or disagree with the information presented [61,62].
The MMSE (Mini Mental State Examination) was used for the cognitive screening of the older adults [63]. This test has the ability to assess five areas of cognition: orientation, registration, attention and calculation, and recall, and language [64,65].
The flowchart (Figure 1) presents the scheme of selection of the older adults according to the sampling, inclusion and exclusion criteria, and screening scales (CES-D, GAI, and MMSE).
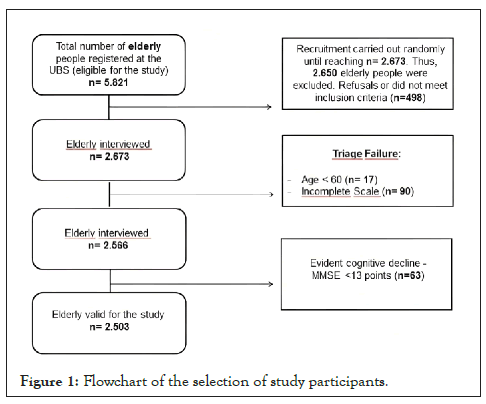
Figure 1: Flowchart of the selection of study participants.
Laboratory methodology
Approximately 5mL (milliliters) of whole blood were collected by venipuncture of a peripheral vein, in tubes containing EDTA anticoagulant (ethylenediamine tetraacetic acid) and were processed and stored in the blood bank of the Laboratório de Patologia Clínica do Instituto de Psiquiatria da Faculdade de Medicina do Hospital das Clínicas (IPq-HC-FMUSP).
We used DNA (Deoxyribo-Nucleic Acid) as the genetic material. DNA extraction was performed using the Salting-out technique [66] and after the extraction we performed purification of the DNA samples. The genetic material was evaluated for its quality and integrity.
DNA genotyping was performed with 27 genetic markers from 11 candidate genes for depression and anxiety disorders. We used the real-time PCR technique for genotypic analysis of polymorphisms, except for the polymorphic variant of the promoter region (5HTTLPR) which we performed by conventional PCR. The table below (Table 1) represents the chosen genes and their respective polymorphisms and alleles.
| Genes | Official name | Polymorphism | Alleles |
|---|---|---|---|
| 5HTT | Solute carrier family 6 member 4 | 5HTTLPR | L/S |
| rs4583306 | A/G | ||
| rs2020933 | A/T | ||
| rs8071667 | C/T | ||
| rs4251417 | C/T | ||
| rs2020942 | C/T | ||
| rs6355 | C/G | ||
| APOE | Apolipoprotein E | rs7412* | C/T |
| rs429358* | C/T | ||
| rs405509 | G/T | ||
| AGTR1 | Angiotensin II receptor type 1 | rs5185 | A/C |
| BDNF | Brain derived neurotrophic factor | rs6265 | C/T |
| COMT | Catechol-O-methyltransferase | rs4680 | A/G |
| rs737865 | A/G | ||
| rs165599 | A/C | ||
| CRP | C-reactive protein | rs1205 | C/T |
| rs3093058 | A/T | ||
| rs1417938 | A/T | ||
| rs1800947 | C/G | ||
| rs3093066 | G/T | ||
| rs2808630 | C/T | ||
| IL6 | Interleukin 6 | rs1800795 | C/G |
| IL10 | Interleukin 10 | rs1800896 | T/C |
| MTHFR | Methylenetetrahydrofolate reductase | rs1801133 | G/A |
| rs1801131 | G/T | ||
| MTOR | Mechanistic target of rapamycin kinase | rs2295080 | G/T |
| TNF | Tumor necrosis factor | rs1800629 | A/G |
Note: *=The combination of the alleles of the rs7412 and rs429358 polymorphisms of the APOE gene results in the alleles e2, e3 and e4.
Table 1: Candidate genes with the 27 polymorphisms and their respective alleles selected for the study.
Statistical analysis
For the analysis in our study, we used the median of the CES-D, GAI, and MMSE scales. The median was calculated from the score obtained in each scale for the 874 older adults. The median value was established in two categorical values; zero (0) or one (1), to indicate a value lower or higher than the median, as shown in detail in Table 2.
| Instrument (scale) | Medians |
|---|---|
| GAI | Median |
| GAI-categorical | Greater than the median (“0”) /Less than the median ("1") |
| MMSE | Median |
| MMSE- categorical | Less than the median (“0”) e Greater than the median ("1") |
| CES-D | Median |
| CES-D - categorical | Greater than the median ("0") e Less than the median ("1") |
Table 2: Categorical values of the median of the applied scales CES-D, GAI, and MMSE.
Using a binary configuration model GAI+2CES-D+4MMSE, Table 3 demonstrates the only possible combinations between the categorical values for the CES-D, GAI, and MMSE scales, thus resulting in eight possible combinations that we denominate Depressive and/or Anxiety Symptomatology Groups (DASG).
| GAI - Cat | MMSE-Cat | CESD-Cat |
|---|---|---|
| 0 | 0 | 0 |
| 1 | 0 | 0 |
| 0 | 0 | 1 |
| 1 | 0 | 1 |
| 0 | 1 | 0 |
| 1 | 1 | 0 |
| 0 | 1 | 1 |
| 1 | 1 | 1 |
Table 3: The possible combinations of categorical values for CES-D, GAI, and MMSE scales.
The Hardy-Weinberg test (Chi square test; p ≤ 0.05) was performed to determine whether the distribution of alleles and their respective genotypes in the population were in balance. In this way, we continue n sequence with the individual and joint analysis of genetic markers.
Genetic markers–statistical analysis
The 27 genetic markers were analyzed in steps. In the first step each polymorphism was analyzed for each of the eight DASG groups. From the genotypic frequencies, the observed allele frequencies were calculated and according to the Hardy-Weinberg Equilibrium (H-W-E) the expected genotypes were calculated. Pearson's chi-square test was performed at a significance level of 5%. When finding an association rejecting H0 at 5%, a chi-square test was performed to compare the rejected group with the other non-rejected groups.
In parallel, we carried out a specific approach to the analysis of APOE polymorphisms (rs429358 and rs7412) for the alleles (ɛ2, ɛ3 e ɛ4), analyzed in relation to the Hardy-Weinberg Equilibrium (H-W-E) by Pearson's chi-square test. The general characteristics between APOE genotypes were described as observed numbers and compared between groups by one-way ANOVA Student´s test. The analysis was performed in SPSS v26 and we considered a statistically significant value of ≤ 0,05.
Joint analysis of genetic markers
The pooled analysis of polymorphisms was performed after the individual analysis. Pearson's chi- square test was also used for genetic markers that showed significant results in the individual analysis. We considered the value of p ≤ 0.05.
In total, 924 blood samples were collected from the 2,503 older people interviewed. Of these collected samples, 50 were excluded, for reasons such as duplicate samples, older people under the age of 60 years, and DNA extractions with poor quality, with a final number of 874 participants.
Genotypic analysis was performed for the 27 polymorphisms. The value of the total number of older people (value of “n”) was different for each genetic marker, as shown in Table 4, because some samples showed uncertain results in the PCR amplification results, and were therefore excluded.
| Gene | Polymorphism | N of valid sample |
|---|---|---|
| 5HTT | 5HTTLPR | 851 |
| 5HTT | rs4583306 | 863 |
| 5HTT | rs2020933 | 858 |
| 5HTT | rs8071667 | 855 |
| 5HTT | rs4251417 | 865 |
| 5HTT | rs2020942 | 865 |
| 5HTT | rs6355 | 867 |
| APOE | rs7412 | 865 |
| APOE | rs429358 | 853 |
| APOE | rs405509 | 852 |
| AGTR1 | rs5186 | 860 |
| BDNF | rs6265 | 862 |
| COMT | rs4680 | 867 |
| COMT | rs737865 | 843 |
| COMT | rs165599 | 865 |
| CRP | rs1205 | 861 |
| CRP | rs3093058 | 863 |
| CRP | rs1417938 | 858 |
| CRP | rs1800947 | 866 |
| CRP | rs3093066 | 864 |
| CRP | rs2808630 | 863 |
| IL6 | rs1800795 | 853 |
| IL10 | rs1800896 | 850 |
| MTHFR | rs1801133 | 865 |
| MTHFR | rs1801131 | 861 |
| MTOR | rs2295080 | 858 |
Table 4: Total valid samples for each genetic marker.
Depressive and/or Anxious Symptomatology Groups (DASG)
For the CES-D, GAI, and MMSE scales, the median value was calculated to establish two categorical values, set at zero (0) and one (1). For CES-D and GAI, we categorized the values above the median as zero (0), and the values below the median as one (1). In contrast, for the MMSE scale, the categorical value zero (0) was adopted for score values below the median, and the categorical value one (1) for values above the median. Table 5 presents the median result and the categorical values for each scale. The association of this categorization is due to the scale scores, since, for both the CES-D and GAI scales, the increase in the score is indicative of depressive and anxious symptomatology, while on the MMSE scale, the decrease in the score indicates lower cognitive capacity.
| Instrument (scale) | Medians |
|---|---|
| GAI | 5 |
| GAI-categorical | >5= 0/ ≤ 5=1 |
| MMSE | 25 |
| MMSE- categorical | <25= 0/ ≥ 25=1 |
| CES-D | 10 |
| CES-D - categorical | >10= 0/ ≤ 10=1 |
Table 5: Categorization of the medians of the GAI, MMSE, and CES-D scales.
Due to the binary configuration model (GAI+2CES-D+4MMSE), we obtained unique combinations of the categorical values of the applied medians, which resulted in eight distinct categories and allowed the construction of the Depressive and/or Anxious Symptomatology Groups of the study (Table 6). The 874 older adults in the study were distributed according to the median scores for each scale (Table 7).
| GAI - Cat | MMSE-Cat | CESD-Cat | DASG |
|---|---|---|---|
| 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 1 |
| 0 | 0 | 1 | 2 |
| 1 | 0 | 1 | 3 |
| 0 | 1 | 0 | 4 |
| 1 | 1 | 0 | 5 |
| 0 | 1 | 1 | 6 |
| 1 | 1 | 1 | 7 |
Table 6: Depressive and/or Anxious Symptomatology Groups (DASG).
| DASG | Total of older adults |
|---|---|
| 0 | 150 |
| 1 | 39 |
| 2 | 41 |
| 3 | 134 |
| 4 | 181 |
| 5 | 65 |
| 6 | 63 |
| 7 | 201 |
| Total | 874 |
Table 7: Total older adults for the eight Depressive and/or Anxious Symptomatology Groups (DASG).
Results of the individual analysis of genetic markers
All 27 polymorphisms were in Hardy-Weinberg equilibrium, so we proceeded with the chi-square analysis. In this individual analysis, we evaluated the possible association between the genotypes of the genetic markers and the DASG, considering statistical significance at the 5% level.
The results of the analysis performed on the distribution of genotypes and the probability of alleles in association with the eight DASG groups that were significant at 5% for a given group (Tables 8-16).
| 5HTT - rs2020933 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Genotype | Expected | Chi-square Calculation | ||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 95 | 49 | 5 | 149 | 0,802 | 95,84 | 47,32 | 5,84 | 149 | 0,007 | 0,060 | 0,121 | 0,188 | 66,45% | Nreject 5% |
| 1 | 32 | 5 | 2 | 39 | 0,885 | 30,52 | 7,96 | 0,52 | 39 | 0,072 | 1,102 | 4,223 | 5,396 | 2,02% | Reject 5% |
| 2 | 21 | 16 | 2 | 39 | 0,744 | 21,56 | 14,87 | 2,56 | 39 | 0,015 | 0,086 | 0,124 | 0,224 | 63,57% | Nreject 5% |
| 3 | 94 | 34 | 2 | 130 | 0,854 | 94,78 | 32,45 | 2,78 | 130 | 0,006 | 0,074 | 0,217 | 0,298 | 58,50% | Nreject 5% |
| 4 | 129 | 49 | 2 | 180 | 0,853 | 130,90 | 45,20 | 3,90 | 180 | 0,028 | 0,320 | 0,927 | 1,274 | 25,90% | Nreject 5% |
| 5 | 44 | 16 | 4 | 64 | 0,813 | 42,25 | 19,50 | 2,25 | 64 | 0,072 | 0,628 | 1,361 | 2,062 | 15,10% | Nreject 5% |
| 6 | 47 | 15 | 0 | 62 | 0,879 | 47,91 | 13,19 | 0,91 | 62 | 0,017 | 0,250 | 0,907 | 1,174 | 27,86% | Nreject 5% |
| 7 | 149 | 43 | 3 | 195 | 0,874 | 149,08 | 42,84 | 3,08 | 195 | 0,000 | 0,001 | 0,002 | 0,003 | 95,93% | Nreject 5% |
| Sum | 611 | 227 | 20 | 858 | 858 | ||||||||||
Table 8: 5HTT gene variant rs2020933: probability of the most frequent allele and distribution of genotypes by DASG group.
| 5HTT- rs8071667 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 93 | 45 | 10 | 148 | 0,780 | 90,14 | 50,73 | 7,14 | 148 | 0,091 | 0,646 | 1,149 | 1,886 | 16,96% | Nreject 5% |
| 1 | 22 | 15 | 2 | 39 | 0,756 | 22,31 | 14,37 | 2,31 | 39 | 0,004 | 0,027 | 0,043 | 0,075 | 78,49% | Nreject 5% |
| 2 | 29 | 6 | 5 | 40 | 0,800 | 25,60 | 12,80 | 1,60 | 40 | 0,452 | 3,613 | 7,225 | 11,289 | 0,08% | Reject 5% |
| 3 | 91 | 35 | 5 | 131 | 0,828 | 89,86 | 37,27 | 3,86 | 131 | 0,014 | 0,138 | 0,334 | 0,486 | 48,56% | Nreject 5% |
| 4 | 117 | 54 | 6 | 177 | 0,814 | 117,15 | 53,69 | 6,15 | 177 | 0,000 | 0,002 | 0,004 | 0,006 | 93,97% | Nreject 5% |
| 5 | 44 | 16 | 4 | 64 | 0,813 | 42,25 | 19,50 | 2,25 | 64 | 0,072 | 0,628 | 1,361 | 2,062 | 15,10% | Nreject 5% |
| 6 | 44 | 18 | 0 | 62 | 0,855 | 45,31 | 15,39 | 1,31 | 62 | 0,038 | 0,444 | 1,306 | 1,788 | 18,12% | Nreject 5% |
| 7 | 133 | 52 | 9 | 194 | 0,820 | 130,31 | 57,37 | 6,31 | 194 | 0,055 | 0,503 | 1,142 | 1,700 | 19,22% | Nreject 5% |
| Sum | 573 | 241 | 41 | 855 | 855 | ||||||||||
Table 9: 5HTT gene variant rs8071667: probability of the most frequent allele and distribution of genotypes by DASG group.
| AGTR1 - rs5186 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 96 | 39 | 12 | 147 | 0,786 | 90,75 | 49,50 | 6,75 | 147 | 0,304 | 2,227 | 4,083 | 6,614 | 1,01% | Reject 5% |
| 1 | 27 | 11 | 1 | 39 | 0,833 | 27,08 | 10,83 | 1,08 | 39 | 0,000 | 0,003 | 0,006 | 0,009 | 92,35% | Nreject 5% |
| 2 | 22 | 17 | 1 | 40 | 0,763 | 23,26 | 14,49 | 2,26 | 40 | 0,068 | 0,436 | 0,699 | 1,203 | 27,27% | Nreject 5% |
| 3 | 80 | 43 | 9 | 132 | 0,769 | 78,05 | 46,91 | 7,05 | 132 | 0,049 | 0,325 | 0,541 | 0,915 | 33,88% | Nreject 5% |
| 4 | 109 | 58 | 13 | 180 | 0,767 | 105,80 | 64,40 | 9,80 | 180 | 0,097 | 0,636 | 1,045 | 1,778 | 18,24% | Nreject 5% |
| 5 | 39 | 23 | 1 | 63 | 0,802 | 40,48 | 20,04 | 2,48 | 63 | 0,054 | 0,437 | 0,883 | 1,375 | 24,10% | Nreject 5% |
| 6 | 40 | 21 | 1 | 62 | 0,815 | 41,13 | 18,73 | 2,13 | 62 | 0,031 | 0,274 | 0,602 | 0,907 | 34,09% | Nreject 5% |
| 7 | 125 | 64 | 8 | 197 | 0,797 | 125,12 | 63,76 | 8,12 | 197 | 0,000 | 0,001 | 0,002 | 0,003 | 95,72% | Nreject 5% |
| Sum | 538 | 276 | 46 | 860 | 860 | ||||||||||
Table 10: AGTR1 gene variant rs5186: probability of the most frequent allele and distribution of genotypes by DASG group.
| BDNF - rs6265 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 110 | 34 | 3 | 147 | 0,864 | 109,72 | 34,56 | 2,72 | 147 | 0,001 | 0,009 | 0,029 | 0,038 | 84,48% | Nreject 5% |
| 1 | 28 | 9 | 2 | 39 | 0,833 | 27,08 | 10,83 | 1,08 | 39 | 0,031 | 0,310 | 0,776 | 1,117 | 29,06% | Nreject 5% |
| 2 | 33 | 8 | 0 | 41 | 0,902 | 33,39 | 7,22 | 0,39 | 41 | 0,005 | 0,084 | 0,390 | 0,479 | 48,88% | Nreject 5% |
| 3 | 94 | 36 | 3 | 133 | 0,842 | 94,32 | 35,37 | 3,32 | 133 | 0,001 | 0,011 | 0,030 | 0,042 | 83,68% | Nreject 5% |
| 4 | 124 | 43 | 11 | 178 | 0,817 | 118,93 | 53,13 | 5,93 | 178 | 0,216 | 1,932 | 4,325 | 6,473 | 1,10% | Reject 5% |
| 5 | 51 | 12 | 1 | 64 | 0,891 | 50,77 | 12,47 | 0,77 | 64 | 0,001 | 0,018 | 0,072 | 0,090 | 76,36% | Nreject 5% |
| 6 | 47 | 14 | 2 | 63 | 0,857 | 46,29 | 15,43 | 1,29 | 63 | 0,011 | 0,132 | 0,397 | 0,540 | 46,24% | Nreject 5% |
| 7 | 142 | 54 | 1 | 197 | 0,858 | 144,98 | 48,04 | 3,98 | 197 | 0,061 | 0,739 | 2,231 | 3,031 | 8,17% | Nreject 5% |
| Sum | 629 | 210 | 23 | 862 | 862 | ||||||||||
Table 11: BDNF gene variant rs6265: probability of the most frequent allele and distribution of genotypes by DASG group.
| COMT - rs165599 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 29 | 75 | 43 | 147 | 0,452 | 30,08 | 72,83 | 44,08 | 147 | 0,039 | 0,064 | 0,027 | 0,130 | 71,83% | Nreject 5% |
| 1 | 11 | 22 | 6 | 39 | 0,564 | 12,41 | 19,18 | 7,41 | 39 | 0,160 | 0,415 | 0,268 | 0,843 | 35,84% | Nreject 5% |
| 2 | 10 | 24 | 6 | 40 | 0,550 | 12,10 | 19,80 | 8,10 | 40 | 0,364 | 0,891 | 0,544 | 1,800 | 17,97% | Nreject 5% |
| 3 | 26 | 78 | 29 | 133 | 0,489 | 31,77 | 66,47 | 34,77 | 133 | 1,047 | 2,001 | 0,957 | 4,005 | 4,54% | Reject 5% |
| 4 | 49 | 91 | 41 | 181 | 0,522 | 49,34 | 90,32 | 41,34 | 181 | 0,002 | 0,005 | 0,003 | 0,010 | 91,97% | Nreject 5% |
| 5 | 22 | 26 | 16 | 64 | 0,547 | 19,14 | 31,72 | 13,14 | 64 | 0,427 | 1,031 | 0,622 | 2,080 | 14,92% | Nreject 5% |
| 6 | 21 | 28 | 14 | 63 | 0,556 | 19,44 | 31,11 | 12,44 | 63 | 0,124 | 0,311 | 0,194 | 0,630 | 42,74% | Nreject 5% |
| 7 | 52 | 83 | 63 | 198 | 0,472 | 44,15 | 98,69 | 55,15 | 198 | 1,395 | 2,496 | 1,117 | 5,007 | 2,52% | Reject 5% |
| Sum | 220 | 427 | 218 | 865 | 865 | ||||||||||
Table 12: COMT gene variant rs165599:probability of the most frequent allele and distribution of genotypes by DASG group.
| CRP - rs1417938 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 2 | 59 | 82 | 143 | 0,220 | 6,94 | 49,12 | 86,94 | 143 | 3,515 | 1,986 | 0,281 | 5,782 | 1,62% | Reject 5% |
| 1 | 2 | 15 | 22 | 39 | 0,244 | 2,31 | 14,37 | 22,31 | 39 | 0,043 | 0,027 | 0,004 | 0,075 | 78,49% | Nreject 5% |
| 2 | 4 | 16 | 20 | 40 | 0,300 | 3,60 | 16,80 | 19,60 | 40 | 0,044 | 0,038 | 0,008 | 0,091 | 76,33% | Nreject 5% |
| 3 | 7 | 48 | 77 | 132 | 0,235 | 7,28 | 47,44 | 77,28 | 132 | 0,011 | 0,007 | 0,001 | 0,018 | 89,20% | Nreject 5% |
| 4 | 11 | 66 | 102 | 179 | 0,246 | 10,82 | 66,37 | 101,82 | 179 | 0,003 | 0,002 | 0,000 | 0,006 | 94,07% | Nreject 5% |
| 5 | 6 | 19 | 39 | 64 | 0,242 | 3,75 | 23,49 | 36,75 | 64 | 1,344 | 0,859 | 0,137 | 2,340 | 12,61% | Nreject 5% |
| 6 | 4 | 27 | 32 | 63 | 0,278 | 4,86 | 25,28 | 32,86 | 63 | 0,153 | 0,117 | 0,023 | 0,292 | 58,87% | Nreject 5% |
| 7 | 16 | 62 | 120 | 198 | 0,237 | 11,16 | 71,69 | 115,16 | 198 | 2,103 | 1,309 | 0,204 | 3,615 | 5,72% | Nreject 5% |
| Sum | 52 | 312 | 494 | 858 | 858 | ||||||||||
Table 13: CRP gene variant rs1417938: probability of the least frequent allele and distribution of genotypes by DASG group.
| IL6 - rs1800795 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 9 | 42 | 92 | 143 | 0,210 | 6,29 | 47,41 | 89,29 | 143 | 1,164 | 0,618 | 0,082 | 1,864 | 17,22% | Nreject 5% |
| 1 | 1 | 13 | 24 | 38 | 0,197 | 1,48 | 12,04 | 24,48 | 38 | 0,156 | 0,077 | 0,009 | 0,242 | 62,29% | Nreject 5% |
| 2 | 1 | 18 | 21 | 40 | 0,250 | 2,50 | 15,00 | 22,50 | 40 | 0,900 | 0,600 | 0,100 | 1,600 | 20,59% | Nreject 5% |
| 3 | 7 | 39 | 87 | 133 | 0,199 | 5,28 | 42,44 | 85,28 | 133 | 0,560 | 0,279 | 0,035 | 0,874 | 34,99% | Nreject 5% |
| 4 | 15 | 49 | 116 | 180 | 0,219 | 8,67 | 61,66 | 109,67 | 180 | 4,625 | 2,601 | 0,366 | 7,592 | 0,59% | Reject 5% |
| 5 | 1 | 20 | 41 | 62 | 0,177 | 1,95 | 18,10 | 41,95 | 62 | 0,464 | 0,200 | 0,022 | 0,686 | 40,76% | Nreject 5% |
| 6 | 7 | 22 | 33 | 62 | 0,290 | 5,23 | 25,55 | 31,23 | 62 | 0,602 | 0,493 | 0,101 | 1,196 | 27,41% | Nreject 5% |
| 7 | 9 | 65 | 121 | 195 | 0,213 | 8,83 | 65,34 | 120,83 | 195 | 0,003 | 0,002 | 0,000 | 0,005 | 94,28% | Nreject 5% |
| Sum | 50 | 268 | 535 | 853 | 853 | ||||||||||
Table 14: IL6 gene variant rs1800795: probability of the least frequent allele and distribution of genotypes by DASG group.
| IL10 - rs1800896 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 22 | 50 | 73 | 145 | 0,324 | 15,23 | 63,53 | 66,23 | 145 | 3,005 | 2,882 | 0,691 | 6,577 | 1,03% | Reject 5% |
| 1 | 2 | 12 | 25 | 39 | 0,205 | 1,64 | 12,72 | 24,64 | 39 | 0,079 | 0,041 | 0,005 | 0,124 | 72,44% | Nreject 5% |
| 2 | 4 | 16 | 19 | 39 | 0,308 | 3,69 | 16,62 | 18,69 | 39 | 0,026 | 0,023 | 0,005 | 0,053 | 81,71% | Nreject 5% |
| 3 | 18 | 52 | 61 | 131 | 0,336 | 14,78 | 58,44 | 57,78 | 131 | 0,702 | 0,710 | 0,180 | 1,592 | 20,70% | Nreject 5% |
| 4 | 22 | 78 | 77 | 177 | 0,345 | 21,02 | 79,95 | 76,02 | 177 | 0,045 | 0,048 | 0,013 | 0,106 | 74,50% | Nreject 5% |
| 5 | 7 | 31 | 26 | 64 | 0,352 | 7,91 | 29,18 | 26,91 | 64 | 0,105 | 0,114 | 0,031 | 0,249 | 61,77% | Nreject 5% |
| 6 | 6 | 28 | 27 | 61 | 0,328 | 6,56 | 26,89 | 27,56 | 61 | 0,047 | 0,046 | 0,011 | 0,105 | 74,61% | Nreject 5% |
| 7 | 26 | 80 | 88 | 194 | 0,340 | 22,45 | 87,09 | 84,45 | 194 | 0,560 | 0,578 | 0,149 | 1,287 | 25,67% | Nreject 5% |
| Sum | 107 | 347 | 396 | 850 | 850 | ||||||||||
Table 15: IL10 gene variant rs1800896: probability of the least frequent allele and distribution of genotypes by DASG group.
| MTOR - rs2295080 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Expected | Chi-square Calculation | |||||||||||||
| Groups | AA | AT | TT | Sum | Allele Probability A | AA | AT | TT | Sum | AA | AT | TT | QUI^2 | p- value | Result |
| 0 | 44 | 62 | 42 | 148 | 0,507 | 38,01 | 73,99 | 36,01 | 148 | 0,945 | 1,942 | 0,998 | 3,885 | 4,87% | Reject 5% |
| 1 | 7 | 16 | 16 | 39 | 0,385 | 5,77 | 18,46 | 14,77 | 39 | 0,263 | 0,328 | 0,103 | 0,693 | 40,50% | Nreject 5% |
| 2 | 11 | 23 | 6 | 40 | 0,563 | 12,66 | 19,69 | 7,66 | 40 | 0,217 | 0,557 | 0,358 | 1,132 | 28,73% | Nreject 5% |
| 3 | 28 | 62 | 41 | 131 | 0,450 | 26,57 | 64,85 | 39,57 | 131 | 0,077 | 0,126 | 0,051 | 0,254 | 61,44% | Nreject 5% |
| 4 | 41 | 87 | 51 | 179 | 0,472 | 39,89 | 89,22 | 49,89 | 179 | 0,031 | 0,055 | 0,025 | 0,111 | 73,91% | Nreject 5% |
| 5 | 14 | 33 | 17 | 64 | 0,477 | 14,54 | 31,93 | 17,54 | 64 | 0,020 | 0,036 | 0,016 | 0,072 | 78,86% | Nreject 5% |
| 6 | 16 | 26 | 19 | 61 | 0,475 | 13,79 | 30,43 | 16,79 | 61 | 0,355 | 0,644 | 0,292 | 1,291 | 25,59% | Nreject 5% |
| 7 | 43 | 89 | 64 | 196 | 0,446 | 39,06 | 96,88 | 60,06 | 196 | 0,397 | 0,640 | 0,258 | 1,295 | 25,51% | Nreject 5% |
| Sum | 204 | 398 | 256 | 858 | 858 | ||||||||||
Table 16: MTOR gene variant rs2295080: probability of the least frequent allele and distribution of genotypes by DASG group.
Some polymorphisms were statistically rejected at 5% for some DASG groups; rs2020933 for group 1 (Table 8); rs8071667 for group 2 (Table 9); rs5186 for group 0 (Table 10); rs6265 for group 4 (Table 11); rs165599 for group 3 and also for group 7 (Table 12); rs1417938 for group 0 (Table 13); rs1800795 for group 4 (Table 14); rs1800896 for group 0 (Table 15); and the polymorphism rs2295080 for group 0 (Table 16).
A new chi-square test was performed for these genetic markers, in which we compared the significant group against the other non-significant groups. The results obtained for the polymorphisms are shown in Tables 17 to 21.
| 5HTT - rs8071667 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Chi-square Calculation | |||||||||||||
| Genotype | CC | CT | TT | Sum | CC | CT | TT | Sum | CC | CT | TT | QUI^2 | Sum | p-value | % |
| Group 2 | 29 | 6 | 5 | 40 | 26,81 | 11,27 | 1,92 | 40 | 0,107 | 2,958 | 3,475 | 6,540 | 6,981 | 0,0305 |
3,05% |
| Complement | 544 | 235 | 36 | 815 | 546,19 | 229,73 | 39,08 | 815 | 0,013 | 0,099 | 0,328 | 0,441 | |||
| Sum | 573 | 241 | 41 | 855 | 855 | ||||||||||
Table 17: 5HTT gene variant rs8071667: significant DASG group (group 2) compared to non-significant group (complement).
| BDNF - rs6265 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Chi-square Calculation | |||||||||||||
| Genotype | CC | CT | TT | Sum | CC | CT | TT | Sum | CC | CT | TT | QUI^2 | Sum | p-value | % |
| Group 2 | 124 | 43 | 11 | 178 | 129,89 | 43,36 | 4,75 | 178 | 0,267 | 0,003 | 8,226 | 8,50 | 6,981 | 0,0305 | 3,05% |
| Complement | 505 | 167 | 12 | 684 | 499,11 | 166,64 | 18,25 | 684 | 0,069 | 0,001 | 2,141 | 2,21 | |||
| Sum | 629 | 210 | 23 | 862 | 862 | ||||||||||
Table 18: BDNF gene variant rs6265: significant DASG group (group 2) compared to non- significant group (complement).
| COMT - rs165599 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Chi-square Calculation | ||||||||||||||
| Genotype | AA | AG | GG | Sum | AA | AG | GG | Sum | AA | AG | GG | QUI^2 | Sum | p-value | % | |
| Group 3 | 26 | 78 | 29 | 133 | 31,34 | 64,69 | 36,97 | 133 | 0,91 | 2,74 | 1,94 | 5,59 | 9,34 | 0,0094 | ||
| Group 7 | 52 | 83 | 63 | 198 | 46,66 | 96,31 | 55,03 | 198 | 0,61 | 1,84 | 1,30 | 3,75 | 0,94% | |||
| Sum | 78 | 161 | 92 | 331 | 331 | |||||||||||
Table 19: COMT gene variant rs165599: comparison between the two significant DASG groups (groups 3 and 7).
| COMT - rs165599 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Chi-square Calculation | |||||||||||||
| Genotype | AA | AG | GG | Sum | AA | AG | GG | Sum | AA | AG | GG | QUI^2 | Sum p-value | % | |
| Group 3 | 26 | 78 | 29 | 133 | 33,827 | 65,65 | 33,519 | 178 | 1,81 | 2,32 | 0,61 | 4,74 | |||
| Group 7 | 52 | 83 | 63 | 198 | 50,358 | 97,74 | 49,901 | 197 | 0,05 | 2,22 | 3,44 | 5,72 | 11,308 | 0,023 | 2,33% |
| Complement | 142 | 266 | 126 | 534 | 135,82 | 263,6 | 134,58 | 487 | 0,28 | 0,02 | 0,55 | 0,85 | |||
| Sum | 220 | 427 | 218 | 865 | 862 | ||||||||||
Table 20: COMT gene variant rs165599: significant DASG groups (groups 3 and 7) compared to non-significant group (complement).
| CRP - rs1417938 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | Chi-square Calculation | |||||||||||||
| Genotype | AA | AG | GG | Sum | AA | AG | GG | Sum | AA | AG | GG | QUI^2 | Sum p-value | % | |
| Group 0 | 2 | 59 | 82 | 143 | 8,67 | 52,00 | 82,33 | 143 | 5,13 | 0,94 | 0,00 | 11,308 | 0,023 | 2,33% | |
| Complement | 50 | 253 | 412 | 715 | 43,33 260,00 411,67 | 715 | 1,03 | 0,19 | 0,00 | 1,21 | |||||
| Sum | 52 | 312 | 494 | 858 | 858 | ||||||||||
Table 21: CRP gene variant rs1417938: significant DASG group (group 0) compared to non-significant group (complement).
As shown in Tables 17 to 21, of the nine polymorphiss that presented results different from the expected p ≤ 0.05 of the groups regarding the distribution of genotypes, only four of these genetic markers presented a statically significant p value when the group with the rejected H0 was compared to the non-rejected groups. Therefore, for our individual analysis, the polymorphisms that presented statistically significant results were: rs8071667 (p=0.03) of the 5HTT gene associated with group 2; rs6265 (p=0.004) of the BDNF gene associated with group 4; rs165599 (p=0.023) of the COMT gene associated with groups 3 and 7; and finally, rs1417938 (p=0.006) of the CRP gene associated with group 0.
For the rs165599 polymorphism in the COMT gene, two groups presented rejected H0, being group 3 and group 7. As shown in Tables 19 and 20, we performed a chi-square test where we placed group 3 against group 7 to test whether they should be aggregated or not in comparison with the other groups. For both tests, the two groups remained associated.
The table below presents the analysis of the rs429358 and rs7412 polymorphisms of the APOE gene both in association with the alleles ɛ2, ɛ3, and ɛ4 and in association with genotypes ɛ2/ɛ2, ɛ2/ɛ3, ɛ2/ɛ4, ɛ3/ɛ3, ɛ3/ɛ4, and ɛ4/ɛ4 (Table 22).
| Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| APOE genotypes | p-value | ||||||||
| 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | ||
| E2/E2 | 0% | 0% | 0% | -1.50% | 0% | 0% | 0% | -0.50% | 0,9852 |
| 17 | 5 | 3 | 14 | 21 | 10 | 6 | 10 | ||
| E2/E3 | -11.90% | -12.80% | -7.50% | -10.50% | -11.70% | -16.10% | -9.50% | -5.20% | 0,6519 |
| 2 | 0 | 2 | 4 | 3 | 1 | 2 | 0 | ||
| E2/E4 | -1.40% | 0% | -5% | -3% | -1.70% | -1.60% | -3.20% | 0% | 0,8981 |
| 88 | 22 | 25 | 78 | 111 | 32 | 41 | 132 | ||
| E3/E3 | -61.50% | -56.40% | -62.50% | -58.60% | -62% | -51.60% | -65.10% | -69.10% | 0,0167 |
| 30 | 12 | 8 | 30 | 41 | 16 | 13 | 43 | ||
| E3/E4 | -21% | -30.80% | -20% | -22.60% | -22.90% | -25.80% | -20.60% | -22.50% | 0,0452 |
| 6 | 0 | 2 | 5 | 3 | 3 | 1 | 5 | ||
| E4/E4 | -4.20% | 0% | -5% | -3.8 | -1.70% | -4.80% | -1.60% | -2.60% | 0,559 |
| APOE alleles | |||||||||
| 19 | 5 | 5 | 22 | 24 | 11 | 8 | 12 | ||
| ɛ2 (T/T) | -6.60% | -6.40% | -6.30% | -8.30% | -6.70% | -8.90% | -6.30% | -3.10% | 0,7989 |
| 223 | 61 | 61 | 200 | 284 | 90 | 101 | 317 | ||
| ɛ3 (T/C) | -78% | -78.20% | -76.30% | -75.20% | -79.30% | -72.60% | -80.20% | -83% | 0,0234 |
| 44 | 12 | 14 | 44 | 50 | 23 | 17 | 53 | ||
| ɛ4 (C/C) | -15.40% | -15.40% | -17.50% | -16.50% | -14% | -18.50% | -13.50% | -13.90% | 0,0491 |
Table 22: APOE (rs429358 and rs7412) genotypes and alleles in association with DASG groups.
Results of the joint analysis of genetic markers
For the joint analysis of the polymorphisms, we used the basis of the individual analysis of each one of them. The analysis was performed by combining the three genotypes of the polymorphisms and performing the general chi-square test for the eight DASG groups, as shown in Table 23.
| COMT - rs165599 + CRP - rs1417938 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Chi-square Calculation | |||||||||||||
| Groups | AA+AA | AA+AT | AA+TT | AG+AA | AG+AT | AG+TT | GG+AA | GG+AT | GG+TT | Sum | QUI^2 | Sum | p- value | % |
| 0 | 31 | 88 | 11 | 77 | 134 | 157 | 45 | 102 | 125 | 770 | 91,08 | |||
| 1 | 13 | 26 | 33 | 24 | 37 | 44 | 8 | 21 | 28 | 234 | 4,13 | |||
| 2 | 14 | 26 | 30 | 28 | 40 | 44 | 10 | 22 | 26 | 240 | 4,87 | |||
| 3 | 33 | 74 | 103 | 85 | 126 | 155 | 36 | 77 | 106 | 795 | 8,36 | |||
| 4 | 60 | 115 | 151 | 102 | 157 | 193 | 52 | 107 | 143 | 1080 | 4,60 | 149,79 | 1,72E-10 | 0,00% |
| 5 | 28 | 41 | 61 | 32 | 45 | 65 | 22 | 35 | 55 | 384 | 10,96 | |||
| 6 | 25 | 48 | 53 | 32 | 55 | 60 | 18 | 41 | 46 | 378 | 6,70 | |||
| 7 | 68 | 114 | 172 | 99 | 145 | 203 | 79 | 125 | 183 | 1188 | 19,08 | |||
| Sum | 272 | 532 | 614 | 479 | 739 | 921 | 270 | 530 | 712 | 5069 | ||||
Table 23: Joint association of polymorphisms rs165599 (COMT) and rs1417938 (CRP).
In the current study, we investigated candidate genes that were reported to be associated with depression and/or anxiety in studies in the literature, that is, genetic markers of susceptibility or vulnerability to the manifestation of these symptoms.
The 27 polymorphisms studied are distributed in 11 distinct genes, namely: 55HTT, APOE, AGTR1, BDNF, COMT, CRP, IL6, IL10, MTHFR, MTOR, and TNF. In Brazil, this study is pioneering in investigating the joint association of candidate gene polymorphisms in a population-based sample of older people. Furthermore, for the analysis of the results, we developed a specific approach based on the median value of the applied scales (CES-D, GAI, and MMSE). We denominated these “Depressive and/or Anxious Symptomatology Groups” (DASG), with the main strategy of this study being the analysis of the association between the investigated polymorphisms and the eight groups formed.
The DASG was a technique deemed necessary to be developed due to the difficulty of practically no Brazilian studies in this area for this specific type of population, and in this way, being able to create references of cut-off points for the CES-D and GAI scales. That is, for the CES-D scale, the majority of international studies use the cut-off value equal to or greater than 16, however, some studies use a lower cut-off point, such as 13 [59], and also while other studies using the highest cut-off point [60]. These differences in the cut-off point for studies using the CES-D are necessary due to different characteristics (socio-economic, educational, and cultural levels) of the samples evaluated. For the Brazilian population, the MMSE scale has different established cut-off points. In addition, several versions of the MMSE adopted the level of education as a criterion to establish the cut-off point [67].
The DASG sought to identify a possible association of polymorphisms and groups subdivided according to the medians of the applied scales. Thus, this DASG classification resulted in eight distinct groups, where we considered group 0 as the worst classification of the medians and group 7 as the best, bearing in mind that, for CES-D and GAI, the increase in the score is indicative of depressive and anxious symptomatology, respectively, while for the MMSE scale the decrease is indicative of cognitive deficit. Through this approach, we were able to better evaluate the results of the investigated genetic markers, that is, to reduce the chances of a false positive. It is important to note that although the older adults in our study with MMSE scores below the median may present mild symptoms of cognitive deficit, there will be no cases of severe symptoms or even dementia, as respondents with an MMSE score below 13 (cut-off point for screening) were excluded.
Of the 27 studied polymorphisms of the 11 candidate genes for depressive and/or anxious symptoms, only four presented statistically significant results at the 5% level (p ≤ 0.05) when analyzed individually. The four polymorphisms were: 5HTT gene rs8071667 for DASG group 2 (p=0.03); BDNF gene rs6265 for group 4 (p=0.004); rs165599 of the COMT gene for groups 3 and 7 (p=0.023) and the rs1417938 polymorphism of the CRP gene for group 0 (p=0.006).
Several studies in the literature observe the possible association with depression of the serotonin transporter gene (5HTT), however, the number of reports of an association involving depression and anxiety is still low [26,68]. A study in 2009 investigated the rs8071667 variant and plasma levels of interleukin-6 with depressive symptoms in twin men aged 54 years, and the lowest frequency allele (T) of this variant was statistically significant p=0.008 [69]. In our study, the same polymorphism was statistically significant p=0.03 associated with group 2 of the DASG, a group consisting of 41 individuals with anxious symptoms and cognition below the median, but without depressive symptoms.
For the BDNF gene, the associated variant rs6265 was also statistically significant in our first analysis, p=0.004, but associated with group 4 of the DASG, a group composed of 181 older people who had symptoms of both depression and anxiety, but without cognitive symptoms (scores above the median for the MMSE scale). There is a report in the literature involving Argentine, American, Brazilian [35,36] and Malay populations associating the A (Met) allele with depression, even though BDNF protein levels are either increased or decreased for these patients [33,36]. Another study in 2018, investigated this variant (rs6265) in depressed patients, and the results showed the presence of the G (Val) allele in most of these participants [35]. Therefore, in the literature we found both the A (Met) and the G allele (Val) associated with the risk of developing depressive disorders.
The rs165599 variant of the COMT gene was also statistically significant for two DASG groups; group 3 (p=0.02), composed of 134 older people who do not have symptoms of depression and anxiety, but have cognition below the median, and group 7 (p=0.02) with a total of 201 individuals who do not present depressive or anxious symptoms and cognition above the median. The COMT gene and its polymorphisms have been studied in psychiatric disorders, including schizophrenia, bipolar disorder, and depression [70,71].
A 2005 study investigated variations in this gene to confer an overall risk for psychiatric disorders and genotyped four polymorphisms, including rs165599 in 394 participants [71]. The G allele of this variant was significantly associated with the diagnoses in all affected individuals, including depression [70]. Despite the few findings in the literature for the rs165599 variant and depression and/or anxiety, the studies found support the idea that this variant may confer a general predisposition to psychiatric disorders.
The rs1417938 polymorphism of the CRP gene was statistically significant (p=0.006) in association with group 0 of the DASG, a group composed of individuals who present depressive and anxious symptoms and also cognition below the median. There are reports in the literature associating depression/anxiety with increased C-reactive protein [72,73].
A study in 2015 investigated 990 older people and genotyped five variants of the CRP gene, among these rs1417938 SNPs. The lowest frequency allele (A) was statistically significant for homozygous women, presenting a reduction in the risk of developing depression when comparing the homozygous women with the most frequent allele [72]. In another finding in the literature, the A allele was potentially associated with the risk for developing depressive disorders [74,75].
Continuing with our findings, the joint analysis was performed based on the results of the individual analysis for each variant. Only two SNPs were significant when analyzed together (rs165599 and rs1417938) of the COMT and CRP genes, respectively. In our analysis, the statistical value of p was 1.72E-10.
As already mentioned, the rs165599 (COMT) variant has been investigated and reported in the literature as a genetic marker that contributes as a potential risk factor for several psychiatric disorders (being reported predominantly for bipolar disorder and schizophrenia) [70]. The rs1417938 polymorphism (CRP) has already been described as an SNP associated with depression [71,72]. The two polymorphisms were statistically significant for the individual analysis and when grouped together, the level of statistical significance was even higher. In this way, it suggests that both variants are an important risk factor for the development of symptoms of depression and anxiety.
For the analysis of APOE (rs429358 and rs7412) we analyzed the SNPs separately (which was not significant in our findings), however, when we performed the genotypic distribution of these two associated polymorphisms for the alleles ɛ2, ɛ3, and ɛ4 and for the genotypes ɛ2/ ɛ2, ɛ2/ ɛ3, ɛ2/ ɛ4, ɛ3/ɛ3, ɛ3/ ɛ4, and ɛ4/ ɛ4 we found statistically significant findings; that is, for the genotype ɛ3/ɛ3 associated with group 0, group 4, and group 7 of the DASG with a p value of 0.016, and for the genotype ɛ3/ɛ4 associated with group 4 and group 7 with a p-value of 0.04. When we analyzed the association only of the alleles, the allele ɛ3 was significant for groups 4 and 7 (p=0.02).
Therefore, the 5HTTLPR variant of the 5HTT gene, which is much reported in the literature, as well as the other investigated genes (AGTR1, IL6, IL10, MTHFR, MTOR, and TNF), showed statistically non-significant results in our sample. Despite reports in the literature associating these genes with depression and/or anxiety, there are also reports corroborating our initial findings.
In the current study, we sought to investigate the relationship between genetic variants of candidate genes and depressive and/or anxious symptoms in a Brazilian population of older people. In addition, we sought to develop an original approach to the analysis by creating DASG to try to partially resolve the lack of information about the study of these polymorphisms in this age group in Brazil. Among the 11 genes studied and their 27 genetic markers, statistically significant results in our sample were observed in only four of them, being rs8071667 (5HTT gene), rs626 (BDNF gene), rs165599 (COMT gene), and rs1417667 (CRP gene). Thus, the most robust results for the joint association of these variants were for the COMT gene variant rs165599 and for the CRP gene variant rs417667.
In the scientific literature, we did not find any other studies with the positive results reported here, however further research is necessary using a similar methodology in larger and independent samples, seeking to reproduce our initial results to confirm the relevance of these findings.
Our study has some methodological limitations. Despite the total sample being composed of a total of 874 older people, the subdivision of the DASG into eight groups based on the CES-D, GAI, and MMSE scores reduces the statistical power to identify true but not detected associations (false negatives), that is, it may make it difficult to detect a positive association for some polymorphisms. In addition to the sample size, the collection of information in our study was carried out by different health professionals, which may be a factor of variability in the approach used and collection of the responses related to each scale used. Furthermore, another relevant point would be the degree of genetic heterogeneity among the individuals studied, which could be a confounding factor in the results presented.
We would like to thank everyone who somehow contributed to the development of the research.
This work was supported by State of São Paulo Research Foundation, Brazil (FAPESP Grants numbers 2012/50010-0 and 2016/07699-8). Isabela Ferreira de Moraes was supported by a Brazilian Federal Research Funding Agency (CAPES-Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – 88882.327657/2019-01).
The present study has no conflicts of interest
The present study has ethical approval.
All study participants signed the Free Consent Form.
Isabela Ferreira de Moraes contributed to the introduction, genotypic analysis and discussion of the article. Thais Chile contributed to the genotypic analysis and discussion of the article. Vanessa de Jesus Rodrigues de Paula contributed to statistical analysis of APOE. Clóvis Alexandrino-Silva contributed to clinical methodology. Geraldo Busatto contributed to clinical methodology. Helena Brentani contributed to statistical analysis of polymorphisms. Homero Vallada contributed to the introduction, discussion and conclusion of the article.
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
Citation: de Moraes IF, de Paula V de JR, Alexandrino-Silva C, Brentani H, Vallada H, Busatto G , et al. (2022) Depression and Anxiety Symptoms in Older Adults: A Joint Association Study of Candidate Genes. J Dep Anxiety.11:486
Received: 19-Sep-2022, Manuscript No. JDA-22-19908; Editor assigned: 23-Sep-2022, Pre QC No. JDA-22-19908 (PQ); Reviewed: 10-Oct-2022, QC No. JDA-22-19908; Revised: 17-Oct-2022, Manuscript No. JDA-22-19908 (R); Published: 24-Oct-2022 , DOI: 10.35248/2167-1044.22.11.486
Copyright: © 2022 de Moraes IF, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.