Research Article - (2021)Volume 7, Issue 8
Description of a Clinical Strain of Mycobacterium cambodiensis sp. nov., a New Member of the Mycobacterium simiae Complex
Fatah Tazerart1,2,3, Jamal Saad1,4, Muriel Militello1,4, Sophie Alexandra Baron1,4, Michel Drancourt1,4* and Sylvain Godreuil5Abstract
A bronchoalveolar lavage sample was collected by bronchoscopy from a 25-year-old Cambodian male patient with suspected clinical tuberculosis and was inoculated in Löwenstein-Jensen medium. Colonies of a rapidly growing, non-chromogenic Gram-positive and acid-fast bacterium were investigated. Scanning electron microscopy showed 1.2 ± 0.29 μm-long and 0.58 ± 0.07 μm-large bacilli that could not be identified using routine matrix-assisted laser desorption ionization-time of flight-mass spectrometry and phenotypic tests (API® ZYM, API® Coryne and Biolog® Phenotype MicroArray assays). In vitro, the isolate was susceptible to isoniazid, amikacin and trimethoprim- sulfamethoxazole. Whole-genome sequencing yielded a 5,703,981-bp draft genome that displayed 69.3% of GC content with 5,207 coding-protein genes and 56 predicted RNA genes, including 3 rRNAs. The rpoB sequence showed 93% sequence similarity with that of Mycobacterium parascrofulaceum in the Mycobacterium simiae complex. Genome sequence-derived DNA-DNA hybridization, OrthoANI and pan-genomic analyses confirmed that this isolate represented an undescribed species within the M. simiae complex. This species was named Mycobacterium cambodiensis after its source of isolation. The isolate was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) with the number CSURP9652.
Keywords
Non-tuberculosis mycobacterium; Mycobacterium simiae complex; Mycobacterium cambodiensis
Introduction
In Cambodia, a country with high tuberculosis burden (incidence, for all clinical forms, estimated at 326 per 100,000 inhabitants in 2017, with 47,000 new cases reported for 16 million inhabitants[1-3], infections caused by non-tuberculous mycobacteria (NTM) also have been recorded. In 2011, the rate of NTM isolation among patients with presumptive multidrug-resistant tuberculosis and positive culture was 26.1% [4]. Among 128 (10.8%) NTM infections diagnosed in patients at Kampong Cham Provincial Reference Hospital between 2012 and 2014, the NTM species could not be identified in 22 cases. Moreover, four patients were co-infected by a NTM and a Mycobacterium tuberculosis complex species [5].
Here, we present the analysis of one isolate, initially referred as strain 716A, from a sputum specimen collected in a Cambodian patient with suspected clinical tuberculosis. We accumulated phenotypically and genetic evidences showing that this isolate belongs to a new species within the Mycobacterium simiae complex. We report the complete polyphasic characterization of strain 716A and its specific features relative to the other known species within the M. simiae complex.
Materials and Methods
Phenotypic characterization
Strain 716A was cultured on Middlebrook 7H10 agar medium (Becton Dickinson, Le Pont de Claix, France) supplemented with 10% OADC (Becton Dickinson) and in the mycobacterial Growth Indicator Tube (MGIT) liquid medium (BACTEC™ MGIT™ 960, Becton Dickinson). Enzyme activities and carbon source utilization were determined by inoculating API® ZYM and API® Coryne strips (bioMérieux, Bruz, France) [6], as described by the manufacturer, with an incubation time of four hours and 24 hours, respectively. The minimum inhibitory concentration (MIC) of the major antimycobacterial agents was determined using the ETEST® (bioMérieux, Craponnes, France).
Biolog phenotype microarray
The capacity of strain 716A to resist or not to 23 inhibitory chemicals and to metabolize 71 different carbon substrates was evaluated using Gen III Microplates Biolog® Phenotype MicroArray (Biolog Inc., Hayward, CA, USA) [7]. Strain 716A was cultured at 37°C on Middlebrook 7H10 agar medium (Becton Dickinson, Franklin Lakes, USA), 10% (v/v) OADC (Becton Dickinson) and 0.5% (v/v) glycerol for two weeks. Colonies were gently taken with sterile swabs, suspended in IF-C2 tubes, and adjusted to 65% transmittance using a turbidimeter (Biolog Inc). Suspensions were then poured into a sterile reservoir and 100 μL of each suspension were deposited in a well of a 96-well plate (with positive and negative control). Two plates (duplicate) were used and incubated in the OmniLog PM System (Biolog Inc.) at 37°C for four days. Results were expressed as area under the curve (AUC) by the Biolog parametric software.
Scanning electron microscopy
The shape and size of strain 716A were determined by scanning electron microscopy (TM4000, Hitachi, Tokyo, Japan) after negative staining at an operating voltage of 15 kV.
MALDI-TOF-MS
The full extraction protocol recommended by Bruker (Bruker Daltonics®, Bremen, Germany) was followed, using glass powder (G8772, Sigma-ALD), pure acetonitrile and formic acid diluted to 70%, as previously described [8]. Then, 1 μL of supernatant was deposited on a ground-steel MALDI target plate. After drying at room temperature, 1 μL of matrix solution (saturated α-cyano-4- hydroxycinnamic acid in 50% acetonitrile, 25% trifluoroacetic acid and 25% H2O) (Sigma-Aldrich) was deposited on the sample. After drying at room temperature, the plate was loaded for analysis in the Microflex LT mass spectrometer (Bruker Daltonics). The results were obtained in the form of spectra and scores; spectra were recorded according to the previously described parameters [9] and were obtained using the MALDI Biotyper (MBT) Compass software, version 4.1.80. The identifications and their scores were obtained with the MALDI Biotyper software, version 4.1.80, and the Mycobacteria Library, version 4.0, database (contains 880 MSPs) (June version, 2017).
DNA preparation and genome sequencing
Total DNA of strain 716A was extracted by vortexing the suspension with glass powder (Sigma- Aldrich, St. Louis, MO, USA) using the FastPrep apparatus (MP Biomedicals, Santa Ana California, USA) and the Qiagen kit (Qiagen, Courtaboeuf, France), as previously described [10]. Then DNA was quantified with the Qubit™ dsDNA HS Assay Kit (Life technologies, Carlsbad, CA, USA), and 0.2 μg/μL of DNA was sequenced with the Illumina MiSeq system (Illumina Inc., San Diego, USA). Paired-end sequencing and automated cluster generation with dual indexed 2× 250-bp reads were performed in a 40-hour run [11].
Genome characterization and genome comparisons
The genome was assembled with SPAdes, version 3.12.0[12], and annotated with Prokka, version 1.13[13]. Emboss GC% and BlastN against the NCBI database were used for detecting the presence of plasmids. The Comprehensive Antibiotic Resistance Database (CARD) was used to identify antibiotic resistance genes. The rpoB gene, TYGS (Type strain genome server) based on the 16S rRNA gene, and Genome BLAST Distance Phylogeny (GBDP) was used to screen all species related to this strain. The similarity among genomes was estimated with the Genome to Genome Distance Calculator (GGDC), available at, and formula 2 was recommended to interpret the results of the analyzed draft genome. The mean levels of relatedness between strain 716A genome sequence and the sequences of other M. simiae complex members were measured with Orthologous Average Nucleotide Identity (OrthoANI). The DDH and OrthoANI values of this strain were calculated with all M. simiae complex members. The Roary pan-genome pipeline in the Galaxy software was used to release the pan-genomic comparison with other Mycobacterium species.
Results
Strain 716A formed orange, smooth and scotochromogenic colonies on Middlebrook 7H10 agar medium, containing 10% (v/v) OADC and 0.5% (v/v) glycerol, after 12-day incubation at 37°C. Biochemical characterization of strain 716A showed a positive reaction to the catalase test, but the oxidase test was negative at room temperature. Investigation of the strain enzymatic activity, using the API ZYM and API CORYNE strips (bioMérieux, Craponne, France), gave positive results for esterase (C4), lipase esterase (C8), lipase (C14), leucine arylamidase, acid phosphatase, naphtol-AS-BI-phosphohydrolase, and alkaline phosphatase. These observations suggested that strain 716A was a Gram-positive bacterium, and Ziehl-Neelsen staining showed pink acid-fast bacilli. Scanning electron microscopy analysis of 100 bacilli indicated that these rod-shaped bacilli measured 1.2 ± 0.29 μm in length and 0.58 ± 0.07 μm in width (Figure 1). The reproducible matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS) profile of strain 716A did not match any of the existing profiles in the Bruker database (version 4.1.80), with an identification score <1.38 (Figure 2). This suggested that strain 716A could be an undescribed species. Strain 716A was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) with the number CSURP9652. Moreover, in vitro antibiotic susceptibility testing showed that strain 716A was susceptible to isoniazid (minimum inhibiting concentration, MIC=0.47 μg/mL), amikacin (MIC=1 μg/mL) and trimethoprim-sulfamethoxazole (MIC=0.19 μg/mL), and resistant to linezolid (MIC>256 μg/ mL), minocycline (MIC>256 μg/mL), doxycycline (MIC>256 μg/mL), rifampicin (MIC>256 μg/mL) and chloramphenicol (MIC>256 μg/mL). Additional phenotypic analyses with the Biolog® Phenotype MicroArray technology (Hayward, California, United States of America) indicated that strain 716A metabolized four carbon sources: Tween 40 (from 24 hours to 7 days), methyl pyruvate and citric acid (from 50 hours to 7 days) and glycerol (only from 90 hours to 7 days). Conversely, strain 716A was not altered by tetrazolium violet during the entire incubation period (from 10 hours to 7 days) (Table 1).

Figure 1: Scanning electron microscopy image of the Mycobacterium cambodiensis strain CSUR9652. Scale bar, 5.00 µm.
Figure 2: MALDI-TOF-MS spectrum of the Mycobacterium cambodiensis strain CSUR9652.
| Position | Substrate | Activity |
|---|---|---|
| D 5 | Glycerol | + |
| F 11 | Tetrazolium Violet | + |
| G 2 | Methyl Pyruvate | + |
| G 5 | Citric Acid | + |
| H 1 | Tween 40 | + |
| A 1 | Negative Control | - |
| A 2 | Dextrin | - |
| A 3 | D-Maltose | - |
| A 4 | D-Trehalose | - |
| A 5 | D-Cellobiose | - |
| A 6 | Gentiobiose | - |
| A 7 | Sucrose | - |
| A 8 | D-Turanose | - |
| A 9 | Stachyose | - |
| A 10 | Positive Control | - |
| A 11 | pH 6 | - |
| A 12 | pH 5 | - |
| B 1 | D-Raffinose | - |
| B 2 | alpha-D-Lactose | - |
| B 3 | D-Melibiose | - |
| B 4 | beta-Methyl-D-glucoside | - |
| B 5 | D-Salicin | - |
| B 6 | N-Acetyl-D-glucosamine | - |
| B 7 | N-Acetyl-beta-D-mannosamine | - |
| B 8 | N-Acetyl-D-galactosamine | - |
| B 9 | N-Acetyl Neuraminic Acid | - |
| B 10 | 1% NaCl | - |
| B 11 | 4% NaCl | - |
| B 12 | 8% NaCl | - |
| C 1 | a-D-Glucose | - |
| C 2 | D-Mannose | - |
| C 3 | D-Fructose | - |
| C 4 | D-Galactose | - |
| C 5 | 3-Methyl Glucose | - |
| C 6 | D-Fucose | - |
| C 7 | L-Fucose | - |
| C 8 | L-Rhamnose | - |
| C 9 | Inosine | - |
| C 10 | 1% Sodium Lactate | - |
| C 11 | Fusidic Acid | - |
| C 12 | D-Serine | - |
| D 1 | D-Sorbitol | - |
| D 2 | D-Mannitol | - |
| D 3 | D-Arabitol | - |
| D 4 | myo-Inositol | - |
| D 6 | D-Glucose-6-PO4 | - |
| D 7 | D-Fructose-6-PO4 | - |
| D 8 | D-Aspartic Acid | - |
| D 9 | D-Serine | - |
| D10 | Troleandomycin | - |
| D11 | Rifamycin SV | - |
| D 12 | Minocycline | - |
| E 1 | Gelatin | - |
| E 2 | Glycyl-L-Proline | - |
| E 3 | L-Alanine | - |
| E 4 | L-Arginine | - |
| E 5 | L-Aspartic Acid | - |
| E 6 | L-Glutamic Acid | - |
| E 7 | L-Histidine | - |
| E 8 | L-Pyroglutamic Acid | - |
| E 9 | L-Serine | - |
| E 10 | Lincomycin | - |
| E 11 | Guanidine HCl | - |
| E 12 | Niaproof 4 | - |
| F 1 | Pectin | - |
| F 2 | D-Galacturonic Acid | - |
| F 3 | L-Galactonic Acid Lactone | - |
| F 4 | D-Gluconic Acid | - |
| F 5 | D-Glucuronic Acid | - |
| F 6 | Glucuronamide | - |
| F 7 | Mucic Acid | - |
| F 8 | Quinic Acid | - |
| F 9 | D-Saccharic Acid | - |
| F 10 | Vancomycin | - |
| F 12 | Tetrazolium Blue | - |
| G 1 | p-Hydroxy-Phenylacetic Acid | - |
| G 3 | D-Lactic Acid Methyl Ester | - |
| G 4 | L-Lactic Acid | - |
| G 6 | lpha-Keto-Glutaric Acid | - |
| G 7 | D-Malic Acid | - |
| G 8 | L-Malic Acid | - |
| G 9 | Bromo-Succinic Acid | - |
| G 10 | Nalidixic Acid | - |
| G 11 | Lithium Chloride | - |
| G 12 | Potassium Tellurite | - |
| H 2 | ama-Amino-Butryric Acid | - |
| H 3 | -Hydroxy-Butyric Acid | - |
| H 4 | β-Hydroxy-D-Lbutyric Acid | - |
| H 5 | Keto-Butyric Acid | - |
| H 6 | Acetoacetic Acid | - |
| H 7 | Propionic Acid | - |
| H 8 | Acetic Acid | - |
| H 9 | Formic Acid | - |
| H 10 | Aztreonam | - |
| H 11 | Sodium Butyrate | - |
| H 12 | Sodium Bromate | - |
Table 1: Scanning electron microscopy image of the Mycobacterium cambodiensis strain CSUR9652. Scale bar, 5.00 µm.
Whole-genome sequencing yielded 128 scaffolds corresponding to 5,703,981 bp with a GC content of 69.3% (GCA_902652685.1). The genome was predicted to encode 5,263 genes, including 5,207 protein-coding genes and 56 RNAs (52 tRNA, three rRNA, and one tmRNA). Nucleotide BLAST (blastn suite) against the standard nucleotide collection (nr/nt) database using Megablast (optimized for highly similar sequences) of the 3,453-bp rpoB gene showed that strain 716A was related to the M. simiae complex, with 93.86% of sequence similarity to the homologous rpoB sequence of Mycobacterium parascrofulaceum. In addition, the 16S rRNA gene sequence indicated that Mycobacterium saskatchewanense DSM 44616, Mycobacterium interjectum ATCC 51457, Mycobacterium paraense IEC26 and Mycobacterium palustreDSM 44572 were the closest species to strain 716A, but with low percentage of identity (26.6%, 25.8%, 25.4% and 25.3%, respectively). In the phylogenetic tree built using Genome BLAST Distance Phylogeny (GBDP), strain 716A was closest to M. saskatchewanense among the 19 members of the M. simiae complex (Figures 3). In silico DNA- DNA hybridization (DDH) analysis of strain 716A yielded less than 30% of identity with the different members of the M. simiae complex (Table 2). Moreover, the overall similarity between the strain 716A sequence and those of M. simiae complex members, measured with the OrthoANI algorithm, ranged between 83.3% and 78.19% (Table 3). Pan-genome analysis of strain 716A within the M. simiae complex yielded 36,451 genes, including 1,103 core genes, 523 soft core genes, 4,941 shell genes shared by several species, and 29,884 cloud genes unique to one species. The pan- genome tree showed that strain 716A was more closely related to M. saskatchewanense (Figures 4).
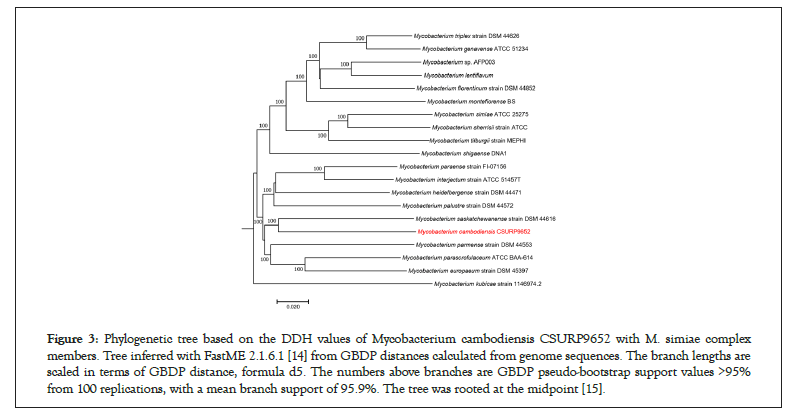
Figure 3: Phylogenetic tree based on the DDH values of Mycobacterium cambodiensis CSURP9652 with M. simiae complex members. Tree inferred with FastME 2.1.6.1 [14] from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance, formula d5. The numbers above branches are GBDP pseudo-bootstrap support values >95% from 100 replications, with a mean branch support of 95.9%. The tree was rooted at the midpoint [15].
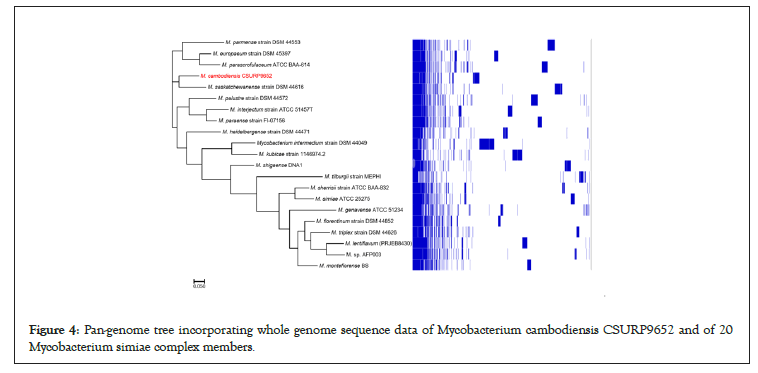
Figure 4: Pan-genome tree incorporating whole genome sequence data of Mycobacterium cambodiensis CSURP9652 and of 20 Mycobacterium simiae complex members.
| Strains | CSURP9652 (DDH %) |
|---|---|
| M. saskatchewanense strain DSM 44616 | 26.6 |
| M. interjectum strain ATCC 51457T | 25.8 |
| M. heidelbergense strain DSM 44471 | 25.8 |
| M. paraense strain FI-07156 | 25.4 |
| M. palustre strain DSM 44572 | 25.3 |
| M. parascrofulaceum ATCC BAA-614 | 25.2 |
| M. interjectum strain DSM 44064 | 25.2 |
| M. europaeum strain DSM 45397 | 25 |
| M. parmense strain DSM 44553 | 24.4 |
| M. triplex strain DSM 44626 | 23.5 |
| M. florentinum strain DSM 44852 | 23.2 |
| M. shigaense DNA1 | 23.2 |
| M. genavense ATCC 51234 | 23.1 |
| M. sp. AFP003 | 22.9 |
| M. lentiflavum | 22.8 |
| M. simiae ATCC 25275 | 22.6 |
| M. sherrisii strain ATCC | 22.6 |
| M. montefiorense BS | 22.5 |
| M. kubicae strain 1146974.2 | 22.2 |
| M. intermedium strain DSM 44049 | 21.6 |
Table 2: Comparison of Mycobacterium cambodiensis CSURP9652 with Mycobacterium simiae complex members using GGDC, formula 2 (DDH estimates based on identities/high-scoring segment pair length).
| Strains | CSURP9652 (OrthoANI %) |
|---|---|
| M. saskatchewanense strain DSM 44616 | 83.3 |
| M. heidelbergense strain DSM 44471 | 82.78 |
| M. paraense strain FI-07156 | 82.51 |
| M. interjectum strain ATCC 51457T | 82.42 |
| M. interjectum strain DSM 44064 | 82.39 |
| M. parascrofulaceum ATCC BAA-614 | 82.31 |
| M. palustre strain DSM 44572 | 82.25 |
| M. europaeum strain DSM 45397 | 81.9 |
| M. parmense strain DSM 44553 | 81.45 |
| M. triplex strain DSM 44626 | 80.48 |
| M. florentinum strain DSM 44852 | 80.42 |
| M. genavense ATCC 51234 | 80.3 |
| M. shigaense DNA1 | 80.27 |
| M. sp. AFP003 | 79.98 |
| M. lentiflavum | 79.81 |
| M. montefiorense BS | 79.75 |
| M. sherrisii strain ATCC | 79.65 |
| M. simiae ATCC 25275 | 79.55 |
| M. kubicae strain 1146974.2 | 79 |
| M. intermedium strain DSM 44049 | 78.19 |
Table 3: OrthoANI values of CSURP9652 with Mycobacterium simiae complex members, calculated by the Orthologous Average Nucleotide Identity tool, version.
| Species | Isolation source | Clinical presentation | Isolation year | Characterization year | Isolation site | Growth | Ref |
|---|---|---|---|---|---|---|---|
| M. simiae | Rhesus macaques | ND | ND | 1965 | Hungary | SGM | [19] |
| M. intermedium | Sputum | Lung disease | ND | 1993 | Germany | SGM | [20] |
| M. interjectum | lymph node | Chronic lymphadenitis | ND | 1993 | Germany | SGM | [21] |
| M. genavense | Blood, bone marrow, liver, | Fever, diarrhea, and | ND | 1993 | Geneva | SGM | [22] |
| spleen, intestine, lymph node | weight loss | ||||||
| M. triplex | Lymph node | ND | ND | 1996 | USA | SGM | [23] |
| M. lentiflavum | sputum, gastric juice, urine | Spondylodiscitis | 1991–1993 | 1996 | USA | SGM | [24] |
| M. heidelbergense | Cervical lymph nodes | Lymphadenitis | ND | 1997 | Germany | SGM | [25] |
| M. kubicae | Respiratory specimen | ND | 1994–1997 | 2000 | USA | SGM | [26] |
| M. palustre | Water | ND | 1993 | 2002 | Finland | SGM | [27] |
| M. montefiorense | Moray eels | Granulomatous skin disease | 2001 | 2003 | USA | SGM | [28] |
| M. parmense | Cervical lymph node | Local swelling of | 1999 | 2004 | Italia | SGM | [29] |
| Submandibular what? | |||||||
| M. sherrisii | Clinical specimen | ND | 1975 | 2004 | USA | SGM | [30] |
| M. saskatchewanense | Sputum and pleural fluid | Bronchiectasis | 2000 | 2004 | Canada | SGM | [31] |
| M. parascrofulaceum | Sputum and bronchoscopy samples | Tuberculosis symptoms except for a | 2002 | 2004 | Canada | SGM | [32] |
| dry cough | |||||||
| M. florentinum | Cervical lymph node | Lymphadenopathy | 1993 | 2005 | Italia | SGM | [33] |
| M. stomatepiae | Stomatepia | Granulomatous lesions in spleen | ND | 2008 | London, UK | SGM | [34] |
| mariae spleen tissue | |||||||
| M. europaeum | Sputum | Cavitary pneumopathy | 1995 | 2011 | Italy, Florence | SGM | [35] |
| M. paraense | Sputum | Respiratory symptoms | ND | 2015 | Brazil | SGM | [36] |
| M. ahvazicum | Sputum | Chronic lung disease | 2009 | 2017 | Iran | SGM | [37] |
| M. terramassiliense, M. rhizamassiliense andM. numidiamassiliense | Tomato plant roots | ND | ND | 2018 | France | SGM | [38] |
| M. tilburgii strain MEPHI | Bone marrow | ND | ND | 2019 | Ireland | Uncultu-red | [39] |
| M. cambodiensis | Sputum | Suspicion of clinical tuberculosis | 2013 | 2019 | Cambodia | SGM | This work |
Note: SGM: Slow Growing Mycobacteria, ND: No Data.
Table 4: Synopsis of the M. simiae complex species characterized since 1965.
Discussion
NTM are ubiquitous environmental bacteria [16] that may act as opportunistic pathogens. Lung infection is the most encountered clinical situation [17], and sometimes mimics pulmonary tuberculosis, like in the patient described in this study [18]. Indeed, strain 716A was isolated from a bronchoalveolar specimen collected from a Cambodian patient with suspected pulmonary tuberculosis. In low-income countries, especially those with endemic tuberculosis, such as Cambodia, lung infections are generally considered to be caused by tuberculosis. Therefore, respiratory diseases caused by other bacteria, including NTM, escape detection and this facilitates their spread in the community.
Whole-genome sequence analyses confirmed that strain 716A was a new member of the M. simiae complex, in agreement with its unique phenotypic characteristics. We named this new species Mycobacterium cambodiensis sp. nov. (cam.bo.di.en’sis N.L. adj. neutr. cambodiensis, of Cambodia, the country where the strain was sampled).
M. simiae was first isolated from Rhesus macaques in Hungary in 1965 [19]. To date, there are 23 species belonging to the M. simiae complex. Eight of these species have been isolated from sputum, five from cervical lymph nodes, three from tomato plant roots, two from fishes, one from blood, one from rhesus macaques, one from water, one from bone marrow and one from an unknown human clinical source (Table 4). Among these species, six have been described after 2011 (one in 2017, three in 2018 and two in 2019), indicating that the M. simiae complex is a rapidly expanding complex of mycobacteria.
Conclusion
In conclusion, our data indicated that M. cambodiensis was a new species belonging to the M. simiae complex. This discovery added a new species to the list of species of the genus Mycobacterium responsible for pulmonary infections. The main aim behind the study to interpret a Clinical strain of Mycobacterium cambodiensis sp. is interesting. The study provides limited new information that Mycobacterium cambodiensis was a new species belonging to the Mycobacterium simiae comple.
Acknowledgements
This study was also supported by the French Government under the Investissements d’Avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), [reference: Méditerranée Infection 10-IAHU-03]. Fatah Tazerart benefits from a PhD grant offered by the Algerian Ministry of Higher Education and Scientific Research under the Exceptional National Program (P.N.E.) and the University of Blida 1. The athors acknowledge Hitachi (Toyo, Japan) for providing the laboratory with electron microscopes used in this study.
Author Contributions
F.T. performed the laboratory manipulations and phenotypic characterization of the new species. J.S. carried out Genome characterization and bio-informatics analysis. F.T. and J.S. wrote the main manuscript text, put tables and figures. F.T., J.S. and M.D. conceived the methodology. M.M. carried out the Biolog analyses, processed the data and made the interpretation on software, S.B. made phenotypic and antibiotic susceptibility testing. S.G. and M.D. supervised the phenotypic, genomic, and genetic characterization of the new species. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Ethics Statement
None required.
Availability of Data and Material
The datasets generated and/or analyzed during the current study are available at GenBank under accession numbers: Mycobacterium cambodiensis (GCA_902652685.1).
References
- WHO. World Health Organization Global Tuberculosis Report, World Health Organization. 2019a.
- Prem K, Pheng SH, Teo AK, Evdokimov K, Nang EE, Hsu LY, et al. Spatial and temporal projections of the prevalence of active tuberculosis in Cambodia. BMJ glob health. 2019; 4(1):e001083.
- WHO. World Health Organization. Tuberculosis Country, Regional and Global Profiles: Cambodia.2019b.
- Khann S, Mao ET, Yadav RP. Non-tuberculosis mycobacteria: Trend of isolation rate and characteristics of NTM in Cambodia during 2011–2013. Int J Mycobacteriology. 2015; 4:39.
- Bonnet M, San KC, Pho Y, Sok C, Dousset JP, Brant W, et al. Nontuberculous mycobacteria infections at a provincial reference hospital, Cambodia. Emerg infect dis. 2017; 23(7):1139-1147.
- Bouzid F, Osman DA, Baptiste E, Delerce J, Hassan MO, Arreh WI, et al. Pulmonary Isolation of Multidrug resistant “Mycobacterium simulans” and Mycobacterium tuberculosis from a patient in the Horn of Africa. Sci rep. 2018; 8(1):1-1.
- Bochner BR. Global phenotypic characterization of bacteria. FEMS microbial rev. 2008; 33(1):191-205.
- Wang J, Chen WF, Li QX. Rapid identification and classification of Mycobacterium spp. using whole-cell protein barcodes with matrix assisted laser desorption ionization time of flight mass spectrometry in comparison with multigene phylogenetic analysis. Anal chim acta. 2012; 716:133-137.
- Zingue D, Flaudrops C, Drancourt M. Direct matrix-assisted laser desorption ionisation time-of-flight mass spectrometry identification of mycobacteria from colonies. Eur J Clin Microbiol Infect Dis. 2016;35: 1983–1987.
- Angelakis E, Roux V, Raoult D, Rolain JM. Real-time PCR strategy and detection of bacterial agents of lymphadenitis. Eur j clin microbiol infect dis. 2009; 28(11):1363-1368.
- Saad J, Drancourt M, Hannan MM, Stapleton PJ, Grandjean Lapierre S. Whole-Genome Sequencing of Mycobacterium tilburgii Strain MEPHI. Microbiol resour announc. 2019; 8(40):e00933-19.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J comput biol. 2012; 19(5):455-477.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30(14):2068-2069.
- Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol biol evol. 2015; 32(10):2798-2800.
- Farris JS. Estimating phylogenetic trees from distance matrices. The American Naturalist. 1972; 106(951): 645–668.
- Friedman DZ, Cervera C, Halloran K, Tyrrell G, Doucette K. Nonâ?Âtuberculous mycobacteria in lung transplant recipients: Prevalence, risk factors, and impact on survival and chronic lung allograft dysfunction. Transpl infect dis. 2020; 22(2):e13229.
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am j respir crit care med. 2007;175(4):367-416.
- Agizew T, Boyd R, Mathebula U, Mathoma A, Basotli J, Serumola C, et al. Outcomes of HIV-positive patients with non-tuberculous mycobacteria positive culture who received anti-tuberculous treatment in Botswana: Implications of using diagnostic algorithms without non-tuberculous mycobacteria. 2020; 15(6):e0234646.
- Karassova VA, Weissfeiler J, Krasznay E. Occurrence of atypical mycobacteria in Macacus rhesus. Acta microbiol Acad Sci Hung. 1965; 12(3):275-282.
- Meier A, Kirschner P, Schröder KH, Wolters J, Kroppenstedt RM, Böttger EC, et al. Mycobacterium intermedium sp. nov. I J Syst Microbiol. 1993; 43(2):204-209.
- Lumb R, Goodwin A, Ratcliff R, Stapledon R, Holland A, Bastian I, et al. Phenotypic and molecular characterization of three clinical isolates of Mycobacterium interjectum. J Clin Microbiol. 1997; 35(11):2782-2785.
- Böttger EC, Hirschel B, Coyle MB. Mycobacterium genavense sp. nov. Int j syst bacteriol 1993;43: 841–843.
- Floyd MM, Guthertz LS, Silcox VA, Duffey PS, Jang Y, Desmond EP, et al. Characterization of an SAV organism and proposal of Mycobacterium triplex sp. nov. J clin microbiol. 1996; 34(12): 2963–2967.
- Springer B, Wu WK, Bodmer T, Haase G, Pfyffer GE, Kroppenstedt RM, et al. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996; 34(5):1100-1107.
- Haas WH, Butler WR, Kirschner P, Plikaytis BB, Coyle MB, Amthor B, et al. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J Clin Microbiol. 1997; 35(12):3203-3209.
- Floyd MM, Gross WM, Bonato DA, Silcox VA, Smithwick RW, Metchock B, et al. Mycobacterium kubicae sp. nov., a slowly growing, scotochromogenic Mycobacterium. Int j syst and evol microbiol. 2000; 50(5):1811-1816.
- Torkko P, Suomalainen S, Iivanainen E, Tortoli E, Suutari M, Seppänen J, et al. Mycobacterium palustre sp. nov., a potentially pathogenic, slowly growing mycobacterium isolated from clinical and veterinary specimens and from Finnish stream waters. Int j syst evol microbiol. 2002; 52(5):1519-1525.
- Levi MH, Bartell J, Gandolfo L, Smole SC, Costa SF, Weiss LM, et al. Characterization of Mycobacterium montefiorense sp. nov., a novel pathogenic mycobacterium from moray eels that is related to Mycobacterium triplex. J Clin Microbiol. 2003; 41(5):2147-2152.
- Fanti F, Tortoli E, Hall L, Roberts GD, Kroppenstedt RM, Dodi I, et al. Mycobacterium parmense sp. nov. Int j syst evol microbiol. 2004; 54(4):1123-1127.
- Selvarangan R, Wu WK, Nguyen TT, Carlson LD, Wallis CK, Stiglich SK, et al. Characterization of a novel group of mycobacteria and proposal of Mycobacterium sherrisii sp. nov. J clin microbiol. 2004; 42(1):52-59.
- Turenne CY, Cook VJ, Burdz TV, Pauls RJ, Thibert L, Wolfe JN, et al. Mycobacterium parascrofulaceum sp. nov., novel slowly growing, scotochromogenic clinical isolates related to Mycobacterium simiae. Int j syst evol microbiol. 2004; 54(5):1543-1551.
- Turenne CY, Thibert L, Williams K, Burdz TV, Cook VJ, Wolfe JN, et al. Mycobacterium saskatchewanense sp. nov., a novel slowly growing scotochromogenic species from human clinical isolates related to Mycobacterium interjectum and Accuprobe-positive for Mycobacterium avium complex. Int j syst evol microbiol. 2004; 54(3):659-667.
- Tortoli E, Rindi L, Goh KS, Katila ML, Mariottini A, Mattei R, et al. Mycobacterium florentinum sp. nov., isolated from humans. Int j syst evol microbiol. 2005; 55(3):1101-1106.
- Pourahmad F, Cervellione F, Thompson KD, Taggart JB, Adams A, Richards RH, et al. Mycobacterium stomatepiae sp. nov., a slowly growing, Non-Chromogenic species isolated from Fish. Int j Syst Evol Microbiol. 2008; 58(12): 2821–2827.
- Tortoli E, Böttger EC, Fabio A, Falsen E, Gitti Z, Grottola A, et al. Mycobacterium europaeum sp. nov., a scotochromogenic species related to the Mycobacterium simiae complex. Int j syst evol microbiol. 2011; 61(7):1606-1611.
- Da Costa AR, Fedrizzi T, Lopes ML, Pecorari M, Da Costa WL, Giacobazzi E, et al. Characterization of 17 strains belonging to the Mycobacterium simiae complex and description of Mycobacterium paraense sp. nov. Int j syst evol microbiol. 2015; 65:656-662.
- Bouam A, Armstrong N, Levasseur A, Drancourt M. Mycobacterium terramassiliense, Mycobacterium rhizamassiliense and Mycobacterium numidiamassiliense sp. nov., three new Mycobacterium simiae complex species cultured from plant roots. Sci rep. 2018; 8(1):1-3.
- Bouam A, Heidarieh P, Shahraki AH, Pourahmad F, Mirsaeidi M, Hashemzadeh M, et al. Mycobacterium ahvazicum sp. nov., the nineteenth species of the Mycobacterium simiae complex. Sci rep. 2018; 8(1):1-2.
- Saad J, Phelippeau M, Khoder M, Lévy M, Musso D, Drancourt M, et al. “Mycobacterium mephinesia”, a Mycobacterium terrae complex species of clinical interest isolated in French Polynesia. Sci rep. 2019; 9(1):1-1.
Author Info
Fatah Tazerart1,2,3, Jamal Saad1,4, Muriel Militello1,4, Sophie Alexandra Baron1,4, Michel Drancourt1,4* and Sylvain Godreuil52Department of Laboratoire d'Agro Biotechnologie et de Nutrition des Zones Semi Arides, Université de Tiaret, Tiaret, Algeria
3Institut des Sciences Vétérinaires- Université de Blida 1, Blida, Algeria
4Aix-Marseille-Univ, Marseille, France
5Laboratoire de Bactériologie, CHU Montpellier, MIVEGEC, IRD, CNRS, Université de Montpllier, France
Citation: Tazerart F, Saad J, Militello M, Baron SA, Drancourt M, Godreuil S, et al. (2021) Description of a Clinical Strain of Mycobacterium cambodiensis sp. nov., a New Member of the Mycobacterium simiae Complex. Appli Microbiol Open Access. 7: 211.
Received: 10-Aug-2021 Accepted: 24-Aug-2021 Published: 31-Aug-2021 , DOI: 10.35248/2471-9315.21.7.211
Copyright: © 2021 Tazerart F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.