Journal of Agricultural Science and Food Research
Open Access
ISSN: 2593-9173
ISSN: 2593-9173
Research Article - (2024)Volume 15, Issue 1
Staphylococcus species are considered the primary and most lethal agents that cause mastitis they are also an important pathogens of public health concern because of their production of enterotoxins which causes staphylococcal food poisoning. A total of 592 quarter milk samples, 30 bulk milk samples and 27 swab samples of milkers hands were examined from 12 farms (both mechanized and small holder farms) in Kaduna as well as in Zaria. The prevalence of subclinical mastitis from positive California Mastitis Test (CMT) (≥ +) was 24.5%. The mean Staphylococcal count was 4.2 log10 cfu/ml. One hundred and three (103) Staphyloccocal isolates that were gram positive and catalase positive were identified biochemically, out of which the identities of 51 was confirmed using the Microgen Microbact Kit (MMK), from this number, 25 selected isolates were tested for enterotoxin production using the Staphylococcal Enterotoxin-Reverse Passive Latex Agglutination (SET-RPLA) kit. From the number tested, 60% were found to produce one or more Staphylococcal Enterotoxins (SEs). Eight (32%) of the isolates produced Staphylococcal Enterotoxin A (SEA), 3 (12%) produced Staphylococcal Enterotoxin B (SEB). None of the isolates produced Staphylococcal Enterotoxin C (SEC) and Staphylococcal Enterotoxin D (SED) but 1 (4%) produced SE (ABC). Polymerase Chain Reaction (PCR) analysis to detect the Enterotoxin genes showed that only 5 SEA genes were present out of the 8 SEA producers and 2 of SEB genes were detected in the 3 SEB producers tested. At the end, it was recommended that consumption of raw, unpasteurized cow milk should be avoided by the people in order to prevent the risk of SE food poisoning.
Enterotoxin; Raw milk; Cow; Staphylococcus species
The traditional dairy farms contribute substantially to the milk supply in the country and significantly to poverty alleviation and reduction of malnutrition, it provides a regular source of household income, food and self-employment particularly to the women folk. However, despite the important role of the industry, farmers continue to experience sub optimal performance of their animals due to disease problems especially mastitis. Yet despite the intensive research on the control of bovine mastitis, it still remains the costliest disease of the dairy animals [1].
Microorganisms, particularly bacteria usually gain entry into milk through the udder of the cow by way of the teat canal. The organisms involved, most of which are saprophytic in the outside environment gain access by their ability to grow a short way up into the milk duct of the teat, causing mastitis of the udder [2].
Although several bacterial pathogens can cause mastitis, the genus Staphylococcus is the primary and probably the most lethal agent because it causes chronic and deep infection in mammary glands that are extremely difficult to be cured [3]. Staphylococcus has been found responsible for more than 80% of the subclinical bovine mastitis which may result in about 300 dollars per year of economic losses per animal [4].
In the cows, the main reservoir of Staphylococcus seems to be the infected quarter and transmissions between cows usually occur during milking [5]. Dairy foods are frequently contaminated with Staphylococcus species and Staphylococcus enterotoxins are ranked as one of the most prevalent worldwide, causing gastroenteritis. Staphylococcus aureus is one of the most common causes of food borne acquired infections causing staphylococcal food poisoning characterized by vomiting and diarrhea after the ingestion of heat stable staphylococcal enterotoxins preformed in food by the enterotoxigenic strains. There are several different serological types of staphylococcal enterotoxins including SEA, SEB, SEC, SED, etc., but enterotoxin A is the most commonly associated with staphylococcal food poisoning. Enterotoxins D, E and H and to a lesser extent B, G and I have also been associated with staphylococcal food poisoning.
A sample size of 592 quarter milk samples was collected (from a proposed 600, as 8 blind teats could not supply milk) from about 150 lactating cows for this study based on the prevalence of 30.0% [6].
Bulk milk samples were collected after all the fresh milk from all the lactating cows in the herd were pooled in to a single container. Swab samples were obtained from human milkers and the cows. Any open wound or skin infection on the body of the cattle and or the milkers was targeted for sampling. Where available, they were first disinfected with a piece of cotton wool soaked in 70% alcohol.
California mastitis test
In order to study the quality of milk, the California Mastitis Test (CMT) was carried out on milk samples of composite milk using the CMT kit.
Five ml of each composite and bulk milk samples were collected, each sample was mixed with the reagent and the test carried out according to the manufacturer’s instruction. The criteria used for scoring were
• 0 (negative)
• +1 (weak positive)
• +2 (distinct positive)
• +3 (strong positive).
In this study, CMT score of 0 was regarded or grouped as having originated from cows free of subclinical mastitis and better quality milk, while CMT result of ≥ +1 was taken as evidence of subclinical mastitis and low quality milk [7].
Enumeration of microorganisms
One ml of the raw milk sample was added in to 9 ml of peptone water (pre enrichment). It was homogenized and incubated at 37°C for 24 h. Thereafter, 0.1 ml of this pre-enriched sample was cultured on to Baird-Parker agar (a selective medium for Staphylococcus species) and incubated at 37°C for another 24 h and observed for staphylococcal colonies.
The inocula from each swab stick was also inoculated on to well prepared, sterilized mannitol salt agar by rubbing/streaking the swabs on to the surface of the agar plates and then incubated at 37°C for 24 h and observed for typical colonies of Staphylococcus [8].
The colonies of suspected Staphylococcus species growing on the plates after the incubation were counted and then recorded as the total bacterial/staphylococcal count. For all the colonies, smear preparation and gram staining was performed to guide the way in the characterization of Staphylococcus, which showed typical gram positive bacteria and coccus in clusters.
Identification of Staphylococci
The phenotypic characterization of all the isolates was carried out using biochemical and serological tests.
The following biochemical tests were performed in order to further confirm the identity of the isolates: catalase, coagulase, thermo nuclease production, haemolytic reaction on blood agar, voges paskauer, sugar fermentation/utilization, mannitol fermentation and clumping factor/protein A production tests.
Confirmation of isolates using the Microgen Microbact Testing (MMT) kit
The test was carried out according to the instruction of the kit manufacturer. Four pure colonies from a 24 h culture were picked and emulsified in a 3 ml of suspending medium to a homogenous suspension using a sterile inoculating loop. With a Pasteur pipette, 10 ml of the bacterial suspension was added to the wells of each test strip and incubated at 37°C for 24 h. After incubation, the result was read and recorded on to the MMT organism I.D. report form provided in the kit. Interpretations of the result were aided by a colour chart provided in the kit. From the results entered on the report form, each block of 3 reactions was converted in to a numeric value, the 3 numbers were added together to form 5 digits of the MMT code, which was then entered in to computer aided software for identification.
Enterotoxin production test
By reverse passive latex agglutination: The isolates were tested for Staphylococcus enterotoxin production using an enterotoxin ABCD detection kit by Reverse Passive Latex Agglutination (RPLA) according to the manufacturer’s instruction.
Three well isolated pure colonies of Staphylococcus species were inoculated into Tryptone Soy Broth and incubated at 37°C for 20 h with shaking. The suspension was centrifuged at 900 g for 20 minutes and the supernatant was retained for assay of toxin content.
A V-bottom microtitre plate was then arranged, such that each row had 8 wells. Each sample needed the use of 5 rows. Twenty-five microlitre of the diluent provided in the kit was added to each well in the 5 rows, 25 μl of the test sample was then added into the first well of each of the rows and a doubling dilution with 25 μl conducted to the seventh well. After this, 25 μl of latex sensitized enterotoxins ABCD was added to row 1, 2, 3 and 4 respectively. To the 5th row was added 25 μl of the latex control.
The plates were rotated to enable mixing of the contents of the wells and incubated at 37°C for 24 h [9]. Agglutination was observed against a black background.
Polymerase chain reaction detection of enterotoxin genes
The molecular typing of the enterotoxin gene of the microbial isolates was done by extracting and subjecting the DNA to Polymerase Chain Reaction (PCR), to detect the presence of the gene for enterotoxin production. The primers used (as presented in the Table 1) were provided by Inqaba Biotec, South Africa. The protocol for PCR technique was carried out as described by 1989 [10]. The PCR master mix was prepared according to specification and the amplification of the DNA was done in a thermal cycler with a pre cycle at 94°C for 5 minutes and a final extension at 72°C for 45 seconds.
After amplification, reactions were analysed on 1% agarose gel by electrophoresis. The ladder used was 1 kb plus ladder in vitro gen with expected base pair of the amplicon around 2,500 bp. The bands were viewed in Ultra-Violet (UV) transilluminator.
Califormia mastitis test
Out of the 592 quarter milk samples screened for mastitis (8 samples were omitted due to blind teats) 145 were CMT positive, giving a prevalence of 24.5%. Between farms, the prevalence of subclinical mastitis ranged from 15.0-61.0% (Table 1).
| California Mastitis Test (CMT) reactions | ||||||||
|---|---|---|---|---|---|---|---|---|
| Farm system/location | Farms | Number of samples | - | + | ++ | +++ | ∑(CMT ≥ +) | Prevalence (%) |
| LMDF Kaduna |
X1 | 98 | 80 | 8 | 6 | 4 | 18 | 18 |
| X2 | 60 | 49 | 6 | 3 | 2 | 11 | 18.3 | |
| SHDF Kaduna |
X3 | 39 | 26 | 13 | 0 | 0 | 13 | 33.3 |
| X4 | 40 | 28 | 8 | 4 | 0 | 12 | 30 | |
| X5 | 18 | 7 | 5 | 5 | 1 | 11 | 61 | |
| X6 | 60 | 51 | 4 | 4 | 1 | 9 | 15 | |
| SHDF Zaria |
Y1 | 40 | 28 | 6 | 2 | 4 | 12 | 30 |
| Y2 | 40 | 33 | 5 | 2 | 0 | 7 | 18 | |
| Y3 | 37 | 17 | 14 | 3 | 3 | 20 | 54 | |
| Y4 | 40 | 31 | 3 | 3 | 3 | 9 | 23 | |
| LMDF Zaria |
Y5 | 80 | 68 | 4 | 7 | 1 | 12 | 15 |
| Y6 | 40 | 29 | 3 | 5 | 3 | 11 | 28 | |
| Total | - | 592 | 447 | 79 | 44 | 22 | 145 | 24.5 |
| % | - | - | 75.5 | 13.4 | 7.4 | 3.7 | 24.5 | - |
Note: (-): Negative; (+): Weak positive; (++): Distinct positive; (+++): Strong positive; LMDF: Large Mechanized Dairy Farm; SHDF: Small Holder Dairy Farm.
Table 1: California mastitis test of quarter milk samples.
Out of the 30 bulk milk samples obtained from 30 herds sampled, 19 (63.0%) were negative to CMT, five (16.7 %) were weakly positive and distinctly positive respectively, while only one (3.3%) was strongly positive to CMT (Table 2).
| Farm location | Farm | Herd number | CMT Scores | |||
|---|---|---|---|---|---|---|
| - | + | ++ | +++ | |||
| LMDF Kaduna | x1 | 5 | 3 | 1 | 1 | 0 |
| x2 | 3 | 2 | 0 | 1 | 0 | |
| x3 | 2 | 1 | 1 | 0 | 0 | |
| SHDF Kaduna | x4 | 2 | 1 | 0 | 1 | 0 |
| x5 | 1 | 0 | 0 | 1 | 0 | |
| x6 | 3 | 3 | 0 | 0 | 0 | |
| SHDF Zaria | y1 | 2 | 1 | 0 | 0 | 1 |
| y2 | 2 | 2 | 0 | 0 | 0 | |
| y3 | 2 | 1 | 1 | 0 | 0 | |
| y4 | 2 | 1 | 1 | 0 | 0 | |
| LMDF Zaria | y5 | 4 | 4 | 0 | 0 | 0 |
| y6 | 2 | 0 | 1 | 1 | 0 | |
| Total | 30 | 19 (63%) | 5 (16.7%) | 5 (16.7%) | 1 (3.3%) | |
Note: LMDF: Large Mechanized Dairy Farm; SHDF: Small Holder Dairy Farm; CMT: California Mastitis Test.
Table 2: California mastitis test of bulk milk samples.
Total staphylococcal count from milk and dairy workers
The mean total Staphylococcus count (log10 cfu/ml) was shown in Table 3. The mean total staphylococcal count ranged from 1.43 ± 0.1 to 6.03 ± 0.20 (log10 cfu/ml). The highest mean count (6.03 ± 0.20 log10 cfu/ml) was recorded in the Large Mechanized Dairy Farm (LMDF) in Zaria. Significant differences existed between the counts at p ≤ 0.05 for all locations. The average mean total staphylococcal count (log10 cfu/ml) was 4.26 ± 0.45 (Table 3).
| Location | N | Mean (± SEM) | |
|---|---|---|---|
| Total staphyloccalcount (log10 cfu/ml) | Mean colony count (log10 cfu/ml) | ||
| LMDF Kaduna | 30 | 4.00 ± 0.12 C | 4.26 ± 0.45 |
| SHDF Kaduna | 47 | 5.87 ± 0.01 B | |
| SHDF Zaria | 51 | 5.97 ± 0.01 B | |
| LMDF Zaria | 23 | 6.03 ± 0.20 A | |
| Bulk milk samples | 8 | 2.23 ± 0.15 D | |
| Dairy Workers | 4 | 1.43 ± 0.15 E | |
Note: SEM: Standard Error Mean; N: Number of CMT positive milk samples tested; cfu: Colony forming units; LMDF; Large Mechanized Dairy Farm; SHDF: Small Holder Dairy Farm; A-E: Staphylococcal enterotoxins.
Table 3: Mean (± SEM) total staphylococcal count of milk samples.
Table 4 showed the association between CMT and staphylococcal count at different sampling points. Spearman’s correlation analysis was used to compare the relationship between CMT and staphylococcal count at different sampling points; moderate to high relationship was observed between CMT and staphylococcal count. The highest association between CMT and staphylococcal count was recorded in swab sample (r=0.71) while the least association was recorded in the bulk sample (r=0.27) (Table 4).
| Farm location | CMT | Staphylococcal count |
|---|---|---|
| LMDF Kaduna | 1 | 0.33* |
| 0.33* | 1 | |
| LMDF Zaria | 1 | 0.57** |
| 0.57** | 1 | |
| SHDF Kaduna | 1 | 0.44* |
| 0.44** | 1 | |
| LMDF Zaria | 1 | 0.68** |
| 0.68** | 1 | |
| Bulk samples | 1 | 0.27* |
| 0.27* | 1 | |
| Swab samples | 1 | 0.71** |
| 0.71** | 1 |
Note: (*): p<0.05; (**): p<0.01; CMT: California Mastitis Test; LMDF: Large Mechanized Dairy Farm; SHDF: Small Holder Dairy Farm.
Table 4: Correlation between California Mastitis Test (CMT) and Staphylococcal count at different sampling points.
Phenotypic characterization of Staphylococcus species
Biochemical characterization of the isolates was further confirmed using the Microgen staph I.D kit. A representation of the result of twenty (20) out of the fifty (50) isolates identified with the kit was shown in Table 5 From the test, 19 (38.0%) were identified as Staphylococcus aureus, 9 (18.0%) were Staphylococcus chromogenes, 2 (4.0%) were Staphylococcus hyicus, another 2 (4.0%) were Staphylococcus epidermidis. Only one (2.0%) was Staphylococcus cohnii, 4(8.0%) were Staphylococcus xylosus and then 4(8.0%) were identified as Staphylococcus intermedius (Table 5).
| Isolate no | Octal code | Identity | Probability (%) |
|---|---|---|---|
| 1 | 77766 | Staphylococcus aureus | 99.64 |
| 2 | 36666 | S. chromogenes | 99.98 |
| 3 | 12446 | S. intermedius | 99.86 |
| 4 | 12466 | S. intermedus | 99.86 |
| 5 | 76676 | S. hyicus | 99.49 |
| 6 | 26740 | S. xylosus | 96.64 |
| 7 | 77746 | S. aureus | 98.08 |
| 8 | 36666 | S. chromogene | 99.98 |
| 9 | 72466 | S. aureus | 99.64 |
| 10 | 77762 | S. aureus | 97.85 |
| 11 | 377772 | S. haemolyticus | 96.65 |
| 12 | 47672 | S. hyicus | 99.94 |
| 13 | 76676 | S. hyicus | 99.49 |
| 14 | 36667 | S. chromogenes | 99.95 |
| 15 | 76662 | S. aureus | 99.26 |
| 16 | 76652 | S. hyicus | 99.99 |
| 17 | 67764 | S. aureus | 100 |
| 18 | 26146 | S. xylosus | 99.8 |
| 19 | 30266 | S. epidermidius | 99.83 |
| 20 | 23606 | S. cohnii | 75.21 |
Table 5: Representative phenotypic identification of Staphylococcus species using the microgen staphylococcal identification kit from milk samples and dairy workers.
Tables 6 and 7 showed the distribution of the identified organisms on farm to farm basis (Tables 6 and 7).
| Isolate | Frequency | Percentage |
|---|---|---|
| Staphylococcus aureus | 19 | 38 |
| S. chromogenes | 9 | 18 |
| S. haemolyticus | 2 | 4 |
| S. hyicus | 9 | 18 |
| S. epidermidis | 2 | 4 |
| S. cohnii | 1 | 2 |
| S. xylosus | 4 | 8 |
| S. intermedius | 4 | 8 |
| Total | 50 | 100 |
Table 6: Frequency of occurrence of Staphylococcus species isolated from mastitic milk samples and dairy workers.
| Source | S. aureus, number isolated | S.chromogenes, number isolated | S.intermedius number isolated | S.hycius, numberisolated | S. xylosus, number isolated | S.haemolyticus, number isolated | S. epidermidis, number isolated | S.cohnii, number isolated |
|---|---|---|---|---|---|---|---|---|
| LMDFK | 2 (4.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| SHDFK | 4 (8.0) | 2 (4.0) | 0 (0.0) | 3 (6.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| SHDFZ | 5 (10.0) | 3 (6.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 2 (4.0) | 0 (0.0) | 0 (0.0) |
| LMDFZ | 6 (12.0) | 2 (4.) | 1 (2.0) | 3 (6.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| BMS | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| DW | 1 (2.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 2 (4.0) | 1 (2.0) |
| Total | 19 (38.0) | 9 (18.0) | 5 (10.0) | 10 (20.6) | 4 (8.0) | 2 (4.0) | 2 (4.0) | 1 (2.0) |
Note: LMDFK: Large Mechanized Dairy Farm Kaduna; SHDFK: Small Holder Dairy Farm Kaduna; SHDFZ: Small Holder Dairy Farm Zaria; LMDFZ: Large Mechanized Dairy Farm Zaria; BMS: Bulk Milk Sample; DW: Dairy Workers.
Table 7: Frequency of occurrence of Staphylococcus species (%) from bovine mastitic milk on the basis of farm locations.
Enterotoxin assay
Staphylococcal Enterotoxin (SE) was detected from 15 out of the 25 selected isolates tested, including a standard strain, using the Reverse Passive Latex Agglutination (RPLA) kit and the enterotoxin types (Table 8).
| S/No | Isolate number | Identity | Enterotoxin | |||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| 1 | ATCC 3865 | Staphylococcus aureus | + | - | + | - |
| 2 | LMDFK 25 | Staphylococcus aureus | + | - | - | - |
| 3 | LMDFK 26 | Staph. chromogenes | - | - | - | - |
| 4 | LMDFK 114 | Staphylococcus aureus | + | - | - | - |
| 5 | SHDFZ 303 | Staph. chromogens | - | - | - | - |
| 6 | SHDFZ 250 | Staphylococcus xylosus | - | - | - | - |
| 7 | SHDFZ 271 | Staphylococcus aureus | - | + | - | - |
| 8 | SHDFZ 273 | Staph haemolyticus | - | - | - | - |
| 9 | SHDFZ 279 | Staphylococcus aureus | + | - | + | - |
| 10 | SHDFZ 289 | Staphylococcus aureus | + | - | - | - |
| 11 | SHDFK 355 | Staphylococcus aureus | - | - | - | - |
| 12 | SHDFK 363 | Staphylococcus hyicus | + | - | - | - |
| 13 | SHDFK 366 | Staph. chromogenes | - | - | - | - |
| 14 | SHDFK 401 | Staphylococcus aureus | + | + | - | - |
| 15 | LMDFZ 504 | Staphylococcus aureus | + | + | - | - |
| 16 | LMDFZ 559 | Staphylococcus aureus | + | - | - | - |
| 17 | LMDFZ 565 | Staphylococcus hyicus | - | - | - | - |
| 18 | LMDFZ 580 | S. intermedius | - | - | - | - |
| 19 | LMDFZ 599 | Staphylococcus aureus | - | + | - | - |
| 20 | BMS 2 | Staphylococcu aureus | + | - | - | - |
| 21 | BMS 4 | S. chromogenes | - | + | - | - |
| 22 | BMS 8 | S. intermedius | - | - | - | - |
| 23 | DW 9 | Staphylococcus xylosus | - | - | - | - |
| 24 | DW 19 | Staph. epidermidis | + | - | - | - |
| 25 | DW 25 | Staphylococcus aureus | + | - | - | - |
Note: LMDFK: Large Mechanized Dairy Farm Kaduna; SHDFK: Small Holder Dairy Farm Kaduna; SHDFZ: Small Holder Dairy Farm Zaria; LMDFZ: Large Mechanized Dairy Farm Zaria; BMS: Bulk Milk Sample; DW: Dairy Workers; (+): Positive; (-): Negative.
Table 8: Enterotoxin production test.
Out of the 15 enterotoxin producers, 8 isolates produced enterotoxin type A (32%), 3 isolates produced enterotoxin type B (12%). None produced enterotoxin type C and D but 1 produced a combination of enterotoxin types A and B (4%), 2 (8%) produced a combination of enterotoxin types A and C while only 1 (4%) also produced a combination of enterotoxin types A, B and C. Ten of the isolates tested (40%) were non enterotoxin producers (Table 9).
| SE production | Number of positive isolates | Percentage (%) |
|---|---|---|
| A | 8 | 32 |
| B | 3 | 12 |
| C | 0 | 0 |
| D | 0 | 0 |
| AB | 1 | 4 |
| AC | 2 | 8 |
| ABC | 1 | 4 |
| NIL | 10 | 40 |
| Total | 25 | 100 |
Note: SE: Staphylococcal Enterotoxin; Each letter represents staphylococcal enterotoxin type.
Table 9: Distribution of staphylococcal enterotoxins production by the isolates.
Polymerase chain reaction
The gel pictures of the PCR determination of the presence of the genes encoding staphylococcal enterotoxins were as shown in plates 1 and 2. Only 5 of the 15 isolates subjected to PCR (33%) were positive for SEA gene and 2 (13%) was positive for the SEB gene. None was positive for SEC and SED (Figures 1 and 2).
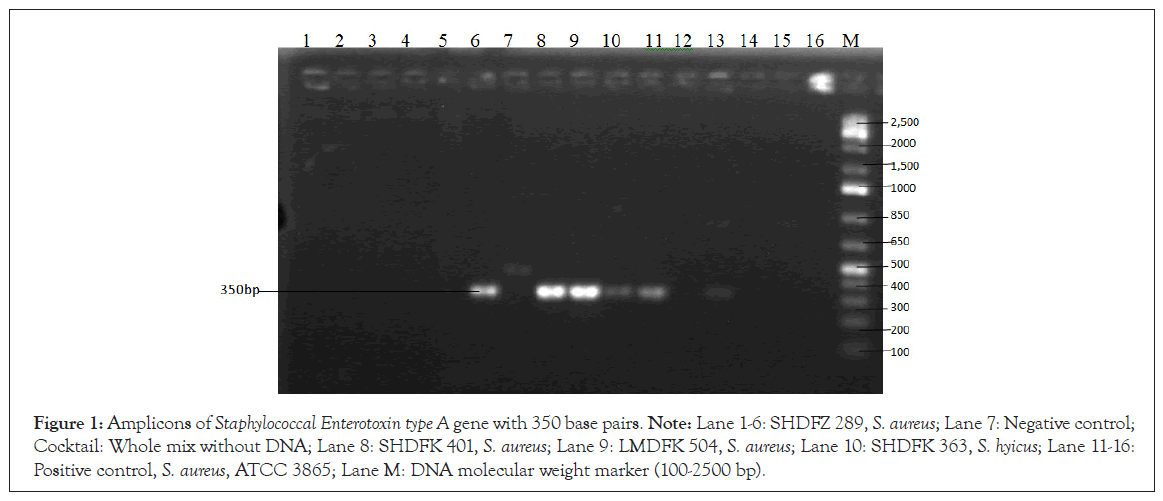
Figure 1: Amplicons of Staphylococcal Enterotoxin type A gene with 350 base pairs. Note: Lane 1-6: SHDFZ 289, S. aureus; Lane 7: Negative control; Cocktail: Whole mix without DNA; Lane 8: SHDFK 401, S. aureus; Lane 9: LMDFK 504, S. aureus; Lane 10: SHDFK 363, S. hyicus; Lane 11-16: Positive control, S. aureus, ATCC 3865; Lane M: DNA molecular weight marker (100-2500 bp).
Figure 2: Amplicon of Staphylococcal Enterotoxin type B (SEB) with 450 base pairs. Note: Lane 1-7: Negative control; Lane 8: SHDFK 401, S. aureus; Lane 9: LMDFZ 504, S. aureus; Lane 11-16: Positive control, S. aureus, ATCC 3865; Lane M: DNA molecular weight marker (100-2,500 bp).
The overall prevalence of mastitis from CMT test in this study was 24.5%. This prevalence is appreciable and may be attributed to the general low level of hygiene observed in the clinical and farm inspection. However, this prevalence is lower compared to 30.5% reported by for traditional dairy herds in Plateau state and 37.0% by in a study carried out in Kaduna and Zaria which is the same study area with this study [11].
The difference could be due to the fact that while the other studies collected milk from nomadic fulani herds only, the present study collected milk from both the traditional small holder farms and the large mechanized dairy farms, whose hygiene measures were higher. Also the sample collection for this study was carried out during the dry season (January to April). This is the period known to record low prevalence of organisms and also the period during which the pH of milk tends to be low, which inhibits the growth of most organisms [12].
However, the result is consistent with the 25.4% reported by in Aydin, Turkey. The prevalence observed in individual farms showed the large mechanized dairy farms to have lower figures than their corresponding small holder dairy farms within the same sampling area. For instance, is was 18.0% and 18.3% in Kaduna large mechanized dairy farms but a prevalence of 30.0- 61.0% was recorded for the small holder farms around Kaduna. This may be attributable to the fact that the large mechanized dairy farms adopted better farm management practices compared to the small holder dairy farms as evidenced in the outcome of farm inspection.
The prevalence of subclinical mastitis observed in the bulk milk samples, 16.7% and 3.3% were in conformity with the reported 15.9% of in Czech Republic in bulk tank milk and the 3.2% reported among nomadic herds by [13]. The lower detection rate of mastitis in the bulk milk samples compared to the quarter milk was probably due to substantial dilution of contaminated milk and this helped to substantially reduce the likelihood of detection as reported by [14].
The bacteriological quality of the fresh raw cow milk samples showed a high total staphylococcal count beyond the standard recommended by the American Public Health Association (APHA, 2001) which is grade A raw milk (<105 cfu/ml) and grade B (milk from local producers) (<106 cfu/ml). The counts for LMDF Kaduna (log10 4.00 cfu/ml), Small Holder Dairy Farm (SHDF) Kaduna (log10 5.87 cfu/ml), SHDF Zaria (log10 5.97 cfu/ml) and LMDF Zaria (log10 6.03 cfu/ml) were all too high, showing that the milk samples were contaminated with bacteria. Only the counts of the bulk milk samples (log10 2.23 cfu/ml) and swab samples (log10 1.43 cfu/ml) are within the standard range.
Counts greater than 103 cfu/ml for raw milk indicates a serious fault in hygiene, the overall mean staphylococcal colony count of log10 4.26 cfu/ml in this study therefore is relatively high and indicative of a milk that has suffered from bacterial contamination. The source of contamination in this study could be attributed to unsatisfactory condition of the housing for the cattle, poor sanitary procedures and or secondary contamination from the skin, mammary gland and nasal cavity of the cows. Contamination could also be from the poor state of health of the milk animals (which could be habouring clinical or subclinical mastitis) and the bacterial causal agents from the udder may get into the milk.
The high level of association observed between CMT and staphylococcal count is not surprising because according to the findings of total bacterial count increase when milk tests positive for mastitis. In the same vein, bacteria that causes mastitis not only contaminate the milk but multiply and grow in the milk due to the fact that the milk is highly nutritious and serves as an excellent growing medium for a wide range of bacteria. Mastitis has been reported as the most significant disease of the dairy industry, causing serious economic losses and species of Staphylococcus especially Staphylococcus aureus was named as one of the most important causative agent all over the world [15]. In the same vein, a study conducted in Egypt that 16% of all mastitic cases were caused by Staphylococcus aureus.
Characterization of the isolates
From the biochemical tests and the subsequent microgen identification, Staphylococcus aureus was the most prevalent organism with 38%. This high detection rate may be due to its contagious nature, which has made it a major udder pathogen in many parts of the world, causing both subclinical and clinical mastitis. This high percentage of Staphylococcus aureus agree with the result that got 34% from cattle in a similar study in Sudan.
The isolation of Staphylococcus aureus is of public health significance since it is a commonly recovered pathogen of food poisoning due to milk and milk products. The other Staphylococcus species (CoNS) detected in this study included Staphylococcus chromogenes (18.0%), Staphylococcus haemolyticus (4.0%), S. hyicus (19.0%), S. epidermidis (4.0%), S. cohnii (2.0%), S.xylosus (8.0%) and Staphylococcus intermedius (8.0%).
This result agreed with that of similar bacteria from bovine mastitis in Iraq. It also agreed with the findings which reported that among researches, isolation of Staphylococcus chromogenes, Staphylococcus epidermidis and S. simulans seem to be the most common Coagulase Negative Staphyclococcus species (CNS) isolated from intra mammary infections inspite of some variations between herds, countries and methods.
Bovine CNS has traditionally been considered as skin flora opportunists and has also been isolated from the cow’s environment [16,17]. Staphylococcus chromogenes was frequently isolated from the teat, skin and teat canal but also from extra mammary sites like nares, hair coat and vagina of cattle [18]. Accordingly Staphylococcus cohnii, S.saprophiticus and S. xylosus were among the most common in the cow’s environment such as in hay and beddings while Staphylococcus haemolyticus is an occasional pathogen of mastitis.
Enterotoxin production
By using a SET-RPLA kit, it was found in this study that the staphylococcal isolates from bovine mastitis in parts of Kaduna state had the ability to produce either Enterotoxin A, SEB, SEA+SEB, SEA+SEC or SEA+SEB+SEC. None produced SED. This result is consistent with previous reports from Japan, Poland and Slovakia. On the other hand, the result was in contrast to other studies carried out in Kenya, Switzerland, Brazil, South Korea and the U.S.A where most enterotoxigenic staphylococcal isolates carried the toxin gene SEC, SEA or SED [19,20].
The most predominant type of staphylococcal enterotoxin produced in this study was SEA. It is known that Staphylococcus enterotoxins are similar in structural and biological properties but differ in amounts produced and in the mechanism of gene regulation. Staphylococcal Enterotoxin A (SEA) is produced throughout the log phase of growth, while Staphylococcal Enterotoxin B (SEB), Staphylococcal Enterotoxin C (SEC) and Staphylococcal Enterotoxin D (SED) are produced in greater quantities during the transition from the exponential to the stationary phase of growth. However, in accordance with the instructions of the manufacturer, SET-RPLA is performed with overnight liquid cultures. This factor might explain why more strains are found positive for SEA using SET-RPLA.
The production of SEA as the most predominant Staphylococcus Enterotoxin (SE) in this study agrees with a review by Genigeorgis, et al., [21] which concluded that the predominant staphylococcal enterotoxin secreted by isolates from food materials and which are involved in staphylococcal gastroenteritis is SEA. But it contrasted with the findings of Tsegmed, et al., [22] who detected only SEC in raw milk from cattle in Mongolia. These differences could be attributed to either difference in techniques used in the studies, differences in origin of the isolates or by geographical differences [23].
Multiple Enterotoxin production was also recorded in this study (SEA+SEB 4%, SEA+SEC 8% and SEA+SEB+SEC 4%). The detection of Staphylococcus species that are able to produce several types of enterotoxins simultaneously has been reported in the past [24,25].
The prevalence of enterotoxin production among the staphylococcal isolates tested in this study was 60%. This is a very high indication that milk can serve as a source of food poisoning by Staphylococcus aureus when not stored properly. But the figure here is even much lower compared to the 74.5% reported in a study conducted on clinical samples in Turkey. This may be due to the fact that human biotypes of Staphylococcus were reported to be enterotoxigenic than animal biotypes.
Molecular characterization
In this study, PCR amplification of the enterotoxin genes revealed that only 4 of the 8 SEA producers and 2 out of the 4 SEB producers actually coded for the genes in PCR amplification thus, showing a lack of correlation between PCR result and the detection of enterotoxin by SET-RPLA. Nonetheless, cases of lack of correlation between SET-RPLA assay and PCR amplification of the gene, has been reported in previous studies [26]. However, one major application of the SET-RPLA assay is the toxin typing of strains for epidemiological purposes, when it is not usually essential to know whether or not a gene is expressed. The occurrence of multiple genes in Staphylococcus species (especially Staphylococcus aureus) is considered rare which explains the negative result obtained for SEC and SED genes in this study [27]. The locations of the enterotoxin genes are also different. SEA is carried on a prophage and SED is carried by a plasmid. The gene for SEC is localized on pathogenicity islands. It is also known that the transcription of SEB, SEC and SED is subject to regulation by the Accessory Gene Regulator (AGR) two-component regulatory system which controls most of the staphylococcal exoprotein virulence factors and that the transcription of SEA is not regulated by the AGR system [28]. SET-RPLA is based on the AGR system, hence more of SEA were produced.
Food-borne intoxication or food poisoning usually occurs after the consumption of food containing the toxin elaborated by the enterotoxigenic strains of Staphylococcus species present as contaminants. Enterotoxins are heat-stable, water soluble proteins that resist most proteolytic enzymes such as pepsin and trypsin and therefore retaining their activity in the digestive tract after ingestion [29].
Most food poisoning strains of Staphylococcus produce enterotoxin A. Although Staphylococcus aureus is the principal Staphylococcus species to cause food poisoning, other Staphylococcus species have also been shown to produce enterotoxins these include Staphylococcus intermedius, S. hyicus, S. xylosus, S.cohnii, S. epidermidis and S. haemolyticus. Although of these, S. intermedius is the only Staphylococcus aureus species to be clearly implicated in food-borne disease.
The following conclusions can be drawn from this study
• There was a high total staphylococcal count (up to 6.03 ± 0.20 log10 cfu/ml) which indicated a high level of milk contamination from unsatisfactory milking practices, this poses a health hazard of food borne infection to consumers through the food chain. The CMT value was about 25%, which is quite appreciable and this equally poses a threat of consumption of mastic milk from consumers and its attendant consequences.
• Different Staphylococcus species were isolated and identified from milk and dairy workers some of which include Staphylococcus chromogenes (18%), S. intermedius (8%), S. haemolyticus (4%) with Staphylococcus aureus being the most prevalent (38%). This is of public health significance because of its association with food poisoning in milk and milk products.
• Staphylococcal enterotoxin production was detected in a good proportion of the isolates tested from the milk samples (up to 60%). This poses a potential public health hazard to consumers of raw milk.
• PCR technique was able to detect the presence of staphylococcal enterotoxin A and B genes (by amplification).
• Dairy farmers should be educated by Government Agricultural Agencies (GAA) and other stakeholders like veterinary and microbiology experts on the need to improve their level of hygiene in milk production and handling, through workshops, seminars and so on.
• Dairy farmers should also be educated on the need to pay greater attention to mastitis control, by employing veterinary services in their animal healthcare from time to time, in order to achieve improved milk yield and quality.
• Consumption of raw, unpasteurized cow milk should be avoided by the people in order to prevent the risk of Staphylococcal enterotoxin food poisoning.
• More research should be intensified in the study of enterotoxigenic Staphylococcus species in milk and other food materials in view of the recent discovery of new Staphylococcus enterotoxins, in order to safe guard the health of consumers.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Umar, Whong, Abdullahi, Aliyu (2024) Detection of Enterotoxin Production by Staphylococcus Species in Fresh Raw Milk Samples from Large Mechanized Dairy Farms and Small Holder Dairy Farms in Kaduna and Zaria. J Agri Sci Food Res. 15:166.
Received: 27-Dec-2023, Manuscript No. JBFBP-23-27393; Editor assigned: 29-Dec-2023, Pre QC No. JBFBP-23-27393 (PQ); Reviewed: 12-Jan-2024, QC No. JBFBP-23-27393; Revised: 19-Jan-2024, Manuscript No. JBFBP-23-27393 (R); Published: 26-Jan-2024 , DOI: 10.35248/2593-9173.24.15.166
Copyright: © 2024 Umar, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.