Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Review Article - (2019)Volume 12, Issue 4
The study of proteomics succeeds the deciphering of the genetic code; this growing burgeoning area is set to dominate scientific research well into the next decade. Current tools employed in studying protein-protein interactions include antibodies, non-protein scaffolds, fluorescence imaging, split enzymes and the relatively new tool termed Adhirons designed by researchers at Leeds University. Antibodies have been used extensively in protein studies due to the high degree of affinity and specificity however the rise in cost and the length of time required to make antibodies has fuelled efforts to find better alternatives. In this work we report whether Adhirons owing to their small size and high stability can be adapted to assay interactions in cells. It will explore whether current tools widely used in protein studies can debunk the davinchi code for protein-protein interactions. Most biological processes are governed by protein interactions and at the heart of most disease states particularly cancer lies a signalling cascade triggered by a plethora of protein interactions. We review current research into proteomics to evaluate and appreciate the work achieved thus far by international scientist crossing east and west divide. The journey into proteomics has already begun and at the present juncture has made significant milestones.
Protein-protein interactions; mRNA; Protein interfaces; Transient complexes
PPIs: Protein-Protein Interactions; QDs: Quantum Dots; FPs: Fluorescent Proteins; Igs: Immunoglobulins
A term which is used sparingly in protein-protein interactions studies is Interactome which denotes the entire number of protein interactions that occur in an organism. The last decade has witnessed resurgence in the study of protein-protein interactions (PPIs). Scientists are now becoming acutely aware of the pivotal role protein interactions play in the smooth regulation of virtually all biological processes in the cell [1,2]. Consequently researchers are now focused on building entire networks of protein interactions, and by finding out the identity of the interaction partners, ultimately leads to the identity of protein function [3,4]. It has been suggested that the total number of protein interactions found in the human genome exceeds 100,000 and to date only a fractions of these interactions have been identified [3]. Further data into interaction studies suggest that a total of 130,000 binary interactions can occur in a human cell at any one time. At this present time, BioGRID (http://thebiogrid.org) a data base for the storage of protein interactions, has listed only 33,943 human protein interactions thus far [3]. Hakes et al. [5] estimates that in yeast approximately 50% of protein interactions have been identified which in contrast to human protein interactions which stands at just 10%. Although the exact number of protein interactions has not been deduced estimates suggest, it is anything from a hundred thousand to around a million [6].
The human genome project identified approximately 30,000 genes, and although this is a major feat for science however, an even greater challenge facing scientists will be to take it a step further, by mapping all the genes and protein interactions. Bonetta et al., Planas-Iglesias et al., Keskin et al. [1,3,7] believes that the challenges of mapping the entirety of protein interactions in the human proteome will be a far greater challenge, than the human genome project, due to the temporal and spatial heterogeneity of the interactome. However, Bonnetta et al. [3] states that exposing pathways, and understanding the role pathways have in disease states, and in the development of disease, is the next mile post in proteomics analysis. There are many reasons why a project of this type will be enormously challenging. One of the caveats presented is alternative splicing. It is estimated that in excess of 90% of all human genes produce alternatively spliced mRNA isoforms. In the human genome, there are approximately 20,000 protein coding genes, of which 196345 multiple transcripts have been released in Ensemble database (GrcH38, version 77), all of this adds to the repertoire of the variety of the human proteome [5,6]. Another complexity is post translational modifications, that all proteins undergo, which include acetylation, phosphorylation, ubiquitination (Figure 1) [8], where a protein is localised in a cell as well as tissue specificity, all add to the complexity of the task [9-11]. A plethora of experimental approaches exist in protein function studies. Many of which have been avidly used in protein identification and protein interaction. These include yeast two-hybrid analysis, Mass spectroscopy and affinity purification to name but a few [3,10]. In addition, complementary bioinformatic approaches have been successfully utlised to identify interactions involving gene clustering and pylogenetic profiling. However, issues around the quality of data suggesting that high numbers of false positives and false negatives throw caution to the accurcy of the data produced (Figure 2) [6]. Although the approaches can identify an interaction, they canot do so in the context of the complex from which the interaction occurs, a key component in understanding function [5,12]. Due in part to limitations in current diagnostic tools this critical area of interactomes, for the most part, remains in the dark. Herein lies the purpose of this study. Adhirons will be adapted as biological cameras using fluorescence microscopy and with the aid of biological techniques such as FRET and BRET. It will bind to a target protein and follow its path along multiple interactions through the cell and with the aid of DNA Paint multiple protein interactions will be visualised in real time. The MAPK pathway is a highly understood pathway and for the purpose of the study, MAPK pathway will demonstrate proof of principle of the utility of Adhirons in protein interaction studies, and that these non-protein scaffolds can be adapted for use in multiple protein interaction studies.

Figure 1: Protein can undergo post-translational modifications, which results in a changes to the protein conformation, by attachment of a phosphate group by specific enzymes. The types of common post-translational modifications are shown in the diagram. Modifications to protein interactions can occur via a number of different pathways. It can result in alterations to the activity of an enzyme, cross talk that is, the same amino acid residue can be changed by more than one type of modification. It can result in alterations to the subcellular localisation of proteins, changes to the ability of protein to bind and the life time of the protein via attachment to different moieties such as ubiquitin Aebersold R [8].
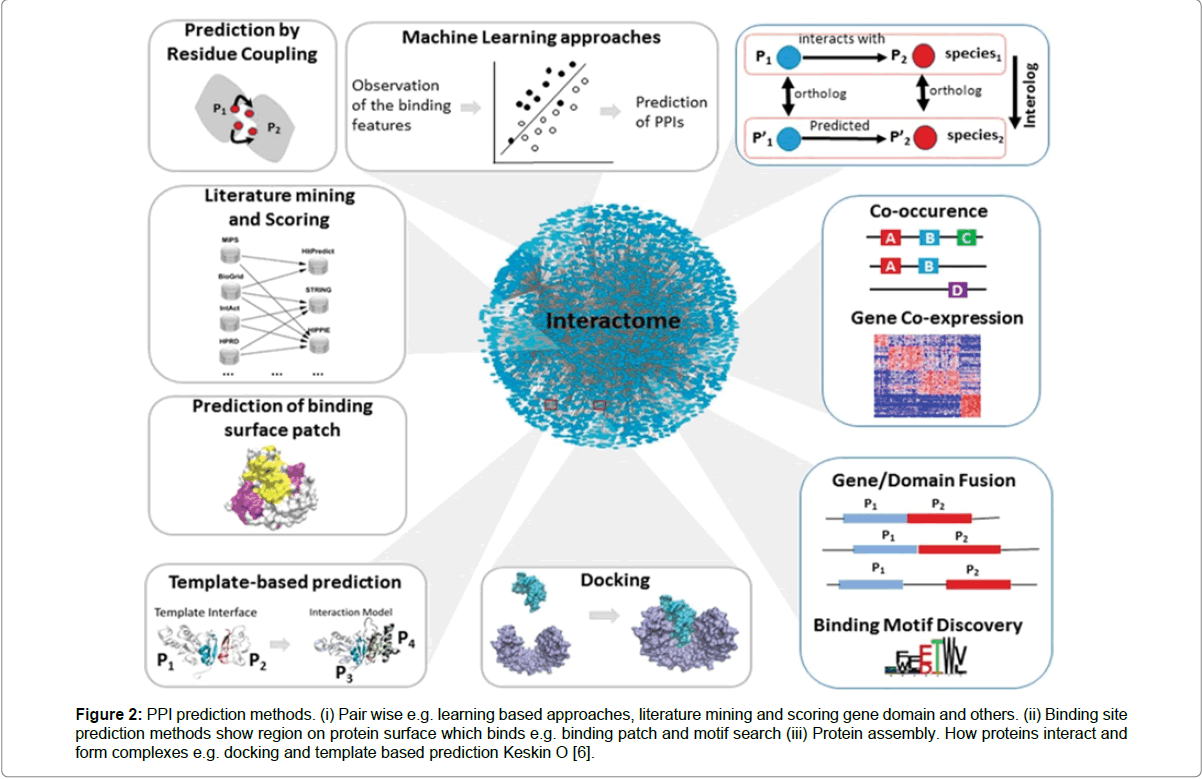
Figure 2: PPI prediction methods. (i) Pair wise e.g. learning based approaches, literature mining and scoring gene domain and others. (ii) Binding site prediction methods show region on protein surface which binds e.g. binding patch and motif search (iii) Protein assembly. How proteins interact and form complexes e.g. docking and template based prediction Keskin O [6].
The Concept of Intrinsic Disorder in Protein Networks
The idea of intrinsic disorder is a concept more commonly found in hub proteins. It has been suggested hub proteins have a higher average of intrinsic disorder in comparison to the average of the rest of the proteome. These protein are typically of two types: transient hub proteins which have a single interface and form multiple transient protein interaction. The second is the multi-interface hubs (Figure 3) which form part of a larger protein complex. The same author Singh et al. suggests that hubs located in protein-protein interactions are divided into two types: “Party” or “Date” hubs. He suggests that intrinsic disorder is evident in party hubs in comparison to date hubs. It is known that hub proteins have larger areas of disordered regions and will tend to have multiple interacting partners and on average have a higher number of disordered residues. The specific resides involved in complexation between two proteins are found located on protein chains on either side of the protein [13]. Binding interactions involve the idea of order and disorder transition, which occurs during transient protein interactions thus enabling reversible interactions. The interactions cause a decrease in conformational entropy resulting in a decrease in affinity thus mediating transient interactions. This is largely due to the lack of molecular contacts occurring in the disordered regions involved in interaction [14]. The localisation of intrinsic disorder in date hubs does allow them to participate in different protein interactions and at different times which is usually observed in signalling pathways. Parts of proteins that fail to fold properly are often in a flexible and unordered state and are described as disordered regions. These regions are regarded as important in protein-protein interactions as they allow for a greater number of protein partners and provide modification sites. About 33% of Eukaryotic proteins are thought to have long disordered regions and this compares to 2-4% of the proteome of archaea and bacteria. Kim et al. further divides the disordered regions into four categories: Molecular recognition, assembly and protein modification as well as entropic chain activity. Furthermore, regions of disordered proteins are known to have functional roles in signalling cascades and an important one is the protein kinase signalling pathway. Intrinsically disordered proteins (IDPs) engage in multiple different protein interactions. It has been suggested that disordered regions are involved in promiscuous binding. The same author (2008) further suggests that the binding partners of single interface hubs are often intrinsically disordered, compared to binding partners of multi-interface hubs. Hence, promiscuous binding occurs via disordered regions in the binding partners and not in the single-interphase hub protein. The propensity of disordered proteins in signalling cascades is believed to be higher. Indeed around 34% of single-interface hubs are kinases and most will target protein kinases (Table 1).
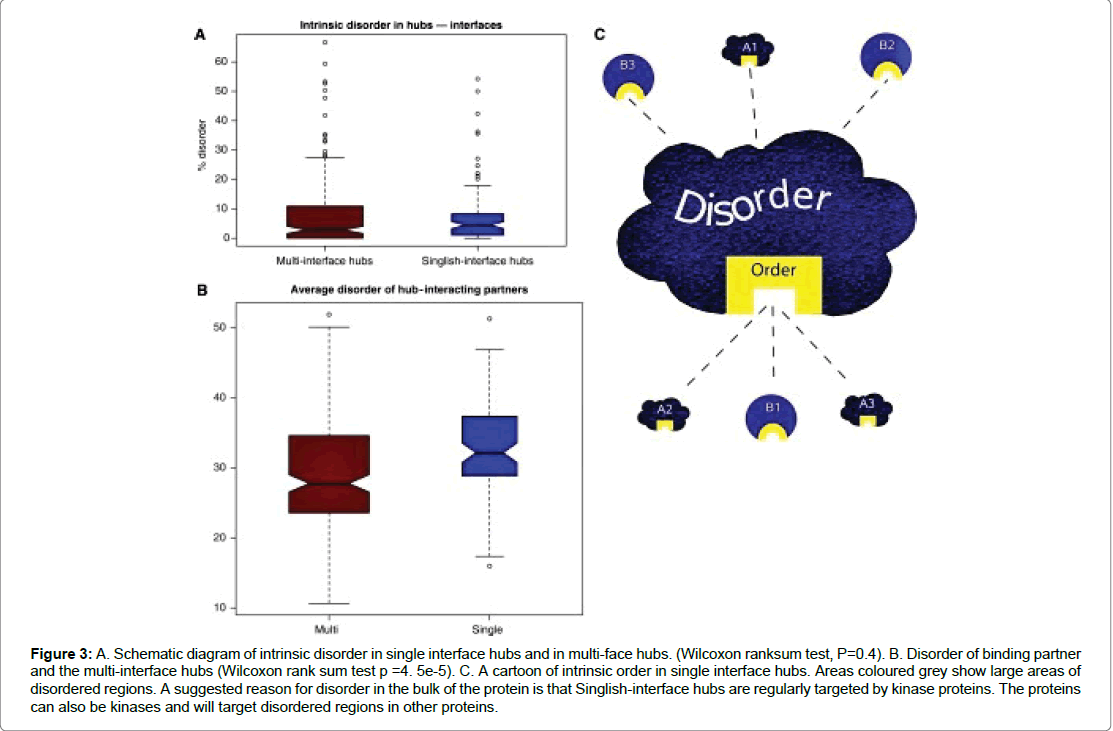
Figure 3: A. Schematic diagram of intrinsic disorder in single interface hubs and in multi-face hubs. (Wilcoxon ranksum test, P=0.4). B. Disorder of binding partner and the multi-interface hubs (Wilcoxon rank sum test p =4. 5e-5). C. A cartoon of intrinsic order in single interface hubs. Areas coloured grey show large areas of disordered regions. A suggested reason for disorder in the bulk of the protein is that Singlish-interface hubs are regularly targeted by kinase proteins. The proteins can also be kinases and will target disordered regions in other proteins.
| Kinase Targets | ||
| Hubs | ||
| Multi-interface | Singlish-Interface | |
| Non-kinase targets | 165 | 56 |
| Kinase targets | 54 | 43 |
| A table of kinase targets versus hub interface. Singlish-interface are targets for kinase proteins (Fisher’s exact test, p=0.001) | ||
Table 1: Table illustrates the binding partners for kinase proteins. Singlish-interface hubs proteins, due to having higher intrinsic disordered regions, are more likely to be targets of kinase proteins. Kinase proteins tend to bind to disordered regions during an interaction as seen in signalling pathways.
Protein Interactions via Protein Interfaces
Protein interfaces mediate protein interactions. These binding sites are specific patches found on both interacting proteins which enable two proteins to complex [15]. Studies have suggested that interactions sites contain chemical and physical recognitions sites that facilitate specific binding between two proteins. Sites of interaction show diversity where some sites can be hydorphobic, planar, globular and others protuding [16]. Generally as recoginition plays a key part in binding, interactions sites are usually found on the surface of proteins located primarily on surface patches [15,16]. Defining surfaces patches is based on Cα atom coordinates that form contiguous circular patches on the surface of proteins [16]. Studies of protein structures reveal that most of the protein interfaces comprise of buried cores surrounded by a rim with a partly accessible rim. On average a typical patch size of an interface will be somewhere between 1600 and 400 Å [15]. Hots spots have been identified through structural analysis studies as mediating almost all of the binding affinity of protein in terfaces. These small and packed regions are highly conserved and have been extensiely studied [15]. Protein interfaces can vary depending on the type of interaction and molecular structure of the interacting proteins. Where an interaction is between domain-peptide complexes around 200-500 Å have been observed. The interface is smaller compared to domaindomain complexes where around 2000 Å of the interface is involved in the interaction [11,13]. Obligate complexes, which account for most homodimers, are observed to be larger in shape and hydrophobic compared to non-obligate complexes, whose residues tend to be polar [9]. Furthermore, residues Leu and Ala occupy interfaces of obligate (permanent) complexes, which are more hydrophobic than the polar residues of non-obligate complexes. Non-obligate complexes are largely made up of residues Ser and Gly [13]. Moreover, PRINT a dataset published by Tuncbag et al. comprised of 8205 interface clusters and each with a different structure. Access to the dataset is available by the following link http://prism.ccbb.ku.edu.tr/. Within the dataset, which stores thousands of protein-protein interactions, 14501 are obligate protein interactions while 2709 are non-obligate protein interactions. Moreover, the Interfaces are grouped into three types: Type 1 where the interface structure and global folds are similar. In Type 2 clusters are of a similar structure but differ in the global folds. Type 3 tend to be multi-partnered and mostly have transient interfaces.
Transient Protein-Protein Interactions
Interfaces have been observed to show conformational changes upon binding to partner protein and are seen to affect interfaces whose size is larger than 1000 Å. An example is the heterotrimeric G protein (Figure 4) [13] where the binding of GTP/GDP exchanges results in the dissociation of Gα and Gβγ subunits. The conformational change triggered by phosphorylation event can cause a big conformational change to the G protein leading to the dissociation of tightly bound complexes [9]. Further, another study found that residues tended to be more conserved in the interfaces than in the rest of the surface of the protein. In addition the study also observed that conservation of central residues was greater than residues found on the periphery of the protein (Table 2) [7]. However, a study by Ofran and Rost [17] raises issues with the results of analysing small data sets such as the study by Jones and Thornton [16]. They argue that analysis of small data set can give results which are far from efficacious but rather that results can be contradictory. In their study they employed a new data mining method which was able to generate the highest data set for interaction studies. It concluded that significant differences existed in amino residues across the different interactions types:
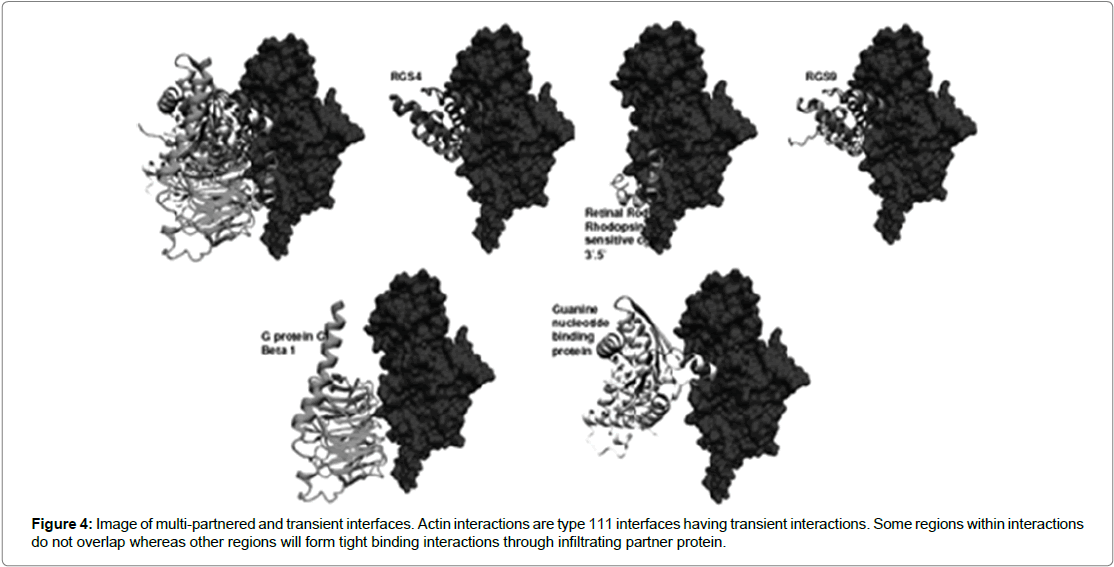
Figure 4: Image of multi-partnered and transient interfaces. Actin interactions are type 111 interfaces having transient interactions. Some regions within interactions do not overlap whereas other regions will form tight binding interactions through infiltrating partner protein.
| Transient\non-obligate | Permanent\obligate | |
| Interface contact area ΔASA (Å2) | <1500 | 1500–10000 |
| Secondary structures | Helix and turns | Helix and β-sheet |
| Interface polarity | High | Low |
| Conformational changes upon binding | Low | High |
| Residue propensity | Polar, charged | Hydrophobic, charged |
| Shape and electrostatic complementarity | High | High |
| Equilibrium dissociation constant (Kd) | >10−6 M (micromolar, μM) | <10−6 M (micromolar, μM) |
Table 2: Structural and kinetic characterization of types of protein–protein complexes.
• Interactions of residues within the same structural domain
• Between different domains
• Between permanent and transient interfaces
• Interactions between homo-oligomers and hetero-oligomers
The differences they conclude were significant that by analysing amino acid composition alone they were able to statistically determine to which of the interface types it belonged within an accuracy of between 63-100%. The results by Ofran and Rost [17] highlight concerns over contradictions obtained by comparison of interfaces. Where some studies reported similarities in the different interfaces between different interaction types and where others studies reported differences. Upon analysis of the characteristic of residues the prevalent residues found in interaction interfaces tended to be polar and charged and contained salt bridges which was the general consensus agreed in most of the studies. Amino acid lysine was found to be minimal in nearly all types of interfaces while arginine was mostly present. Hydrophobic residues histidine, methionine and tyrosine were present in nearly all hydrophobic interfaces contrasted by low amounts of serine, alanine and glycine on the same interface. Findings also suggested that hydrophobic residues were abundant in homooligomers than in hetero-complexes. The results generally agreed with findings of most studies including the study by Jones and Thornton (1993). A study by Ezhurdia et al. raised limitations stating that the paucity of proteins in complexes and the lack of transient complexes, that have yet to be crystalized, and are currently available in The Brookhaven Data Bank (PDB) databases present challenges. It results in a lack of clarity in being able to predict surface residues for the different interaction types. Added to this problem is the time and cost involved in experimentally determining protein-protein complexes. Predicting interface composition for multiple protein interactions will have wide ranging benefits. It will facilitate the development of new drugs and therapies bespoke for a specific protein-protein interaction and will enable the design of mutants to verify protein interactions as occurring (Figure 5) [9].
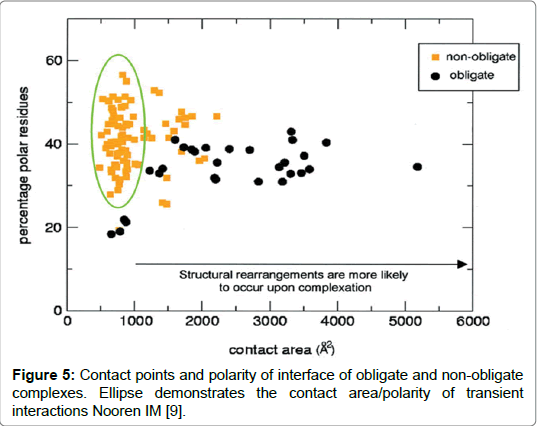
Figure 5: Contact points and polarity of interface of obligate and non-obligate complexes. Ellipse demonstrates the contact area/polarity of transient interactions Nooren IM [9].
Homo and Hetero-oligomer Complexes
A home-oligomer is a protein complex that comprises identical protein units and where a protein interaction occurs involving identical chains it is described as a homo-oligomer. Converse is true for hetero-oligomers as these complexes are formed via interactions occurring between non-identical chains (Figure 6) [9,13,15,16]. Both complexes are made from two components and the characteristic feature, distinguishing each complex, is that homo-complexes provide a scaffold, which can have a more permanent and optimised structure and thus enable stable interactions. Alternatively the hetero-complex transient or non-obligatory transactions are susceptible to breakage but equally can be made depending on the environment [16]. An example is cytochrome c which is a homo-complex whereas, the complex formed from enzyme inhibitor trypsin and the inhibitor bitter gourd, is an example of a hetero-complex (Figure 6) [15,16].
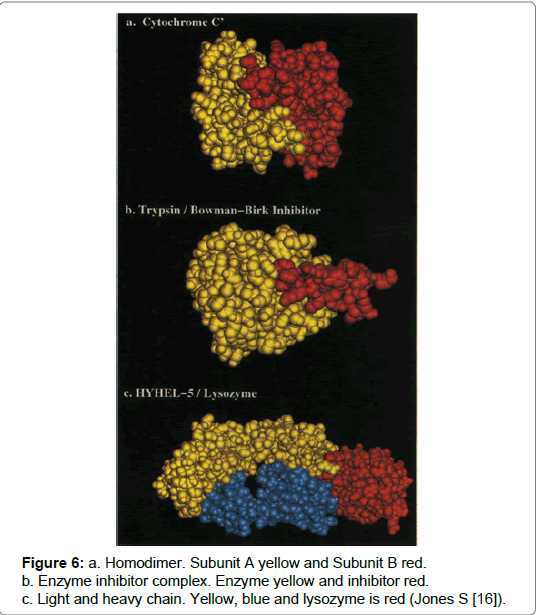
Figure 6: a. Homodimer. Subunit A yellow and Subunit B red.
b. Enzyme inhibitor complex. Enzyme yellow and inhibitor red.
c. Light and heavy chain. Yellow, blue and lysozyme is red (Jones S [16]).
A study by Jones and Thornton [16] analysed the interactions of hetero-complexes and homo-complexes, residues were examined that were commonly associated with the interfaces of both types of complexes. The study found that a high degree of residues were hydrophobic, especially for the homodimer complex, and this was balanced by more polar residues in the hetero-complex. Further, the study revealed that homodimers tended to be more strongly in proteinprotein interactions and were rarely found to exist in a monomer state Ezkudia et al. Further structural studies into PPIs suggest interfaces in the majority of homodimers were observed to be larger and hydrophobic in composition. Secondly, stable complexes could be formed with co-protomers via large and intertwined interfaces in comparison to hetero-complexes that had a more polar composition at the interface [9]. Further, results concluded that hydrophobicity was observed to be the greatest in homo-dimers which concurred with previous studies. For hetero-complexes on the parameter of solvation (the potential is a measure of preference for burial or exposure to solvent) no trend was observed and neither for parameter of hydrophobicity. Of the proteins studied in the hetero-complex, where the hetero-complex could also exist in the monomeric and non-complexed form, the interfaces observed were less hydrophobic than proteins in a complexed state. The polarity depended largely on the function of the protein (Figure 7). For the Barnase the interface was found to be polar this was due to its function. Barnase is a ribonuclease and it binds to RNA.
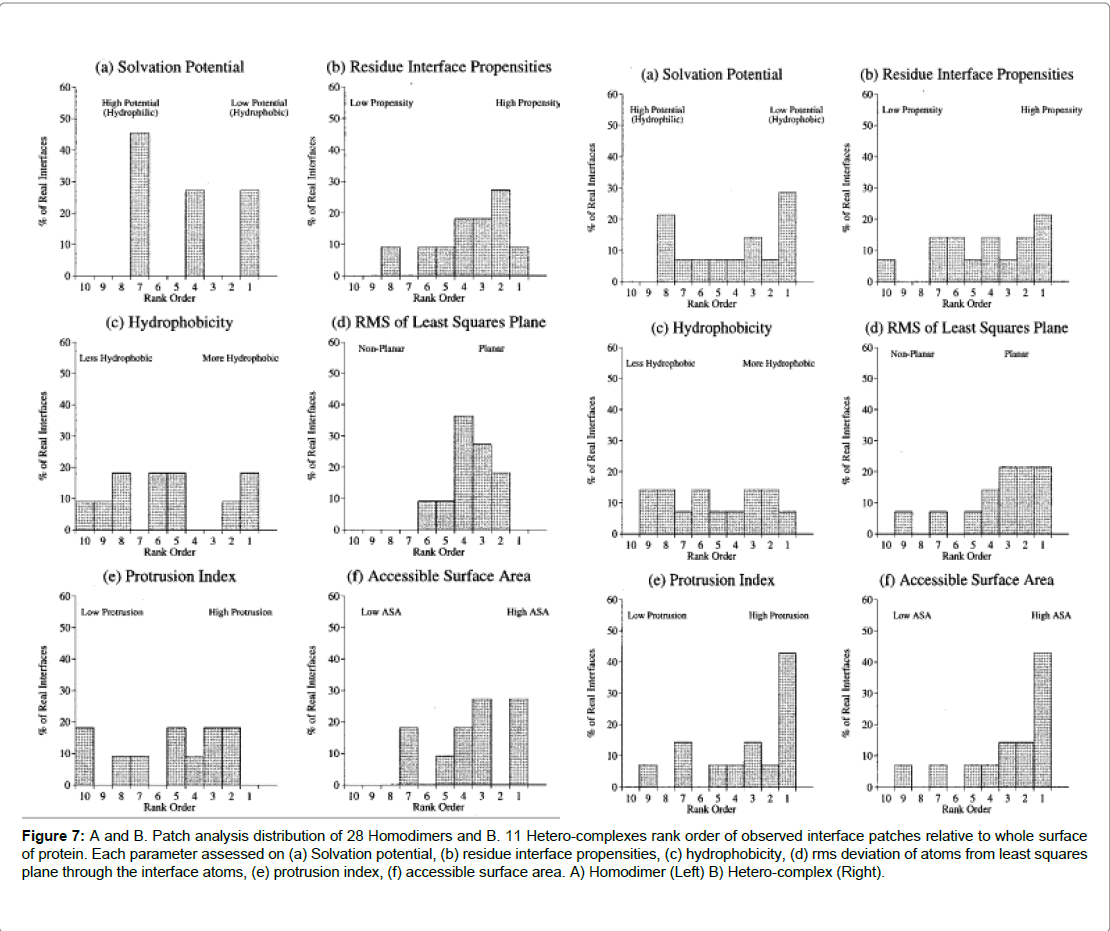
Figure 7: A and B. Patch analysis distribution of 28 Homodimers and B. 11 Hetero-complexes rank order of observed interface patches relative to whole surface of protein. Each parameter assessed on (a) Solvation potential, (b) residue interface propensities, (c) hydrophobicity, (d) rms deviation of atoms from least squares plane through the interface atoms, (e) protrusion index, (f) accessible surface area. A) Homodimer (Left) B) Hetero-complex (Right).
Obligate and Non-Obligate Complexes
The main difference between obligate and non-obligate complexes centres on whether the protomers are mutually independent of each other. Azbabacan et al. states that where complexes which were made up of protomers and monomers, were unstable in vivo, is characteristic of an obligate interaction. The complexes also differed in their biological role where the majority of homodimers existed in their multimeric state and only by denaturing the individual monomers was it possible to separate them [16]. Secondly, interaction occurred via identical chains, in the case of homo-oligomer and non-identical chains in the case of a hetero-complex [9]. Polar residues lined the interfaces of non-obligate interactions and had a characteristically smaller interface size. Therefore, resulting in interactions that were less stable and more transient in their binding nature. However, obligate complexes had a more permanent interaction. Another difference observed by Keskin et al. [7] was in the difference between the two complexes with regards to the relative contribution to the physical interaction. The study found that in obligate complexes, interactions were generally water tight with hardly any water molecules trapped between the monomers. A strong hydrophobic interaction was also deduced with a high degree of complementarity between interfaces of partner proteins. Transient complexes associated with non-obligate complexes were smaller and had a higher number of polar/charged residues. It was observed that in the interface, a distinct absence of optimization existed between interacting proteins, which consequently caused weak and transient binding interactions. Secondly, obligate complexes had residues which evolved quite slowly thus enabling the protein and partner protein to coevolve within a complex. This is in contrast to the interfaces of permanent interactions with tight binding showed an increase rate of mutations at the interface with no correlated mutations.
Transient and Permanent Complexes
Transient complexes play important roles in regulating chemical and signalling pathways in cells. A number of key processes which occur in the cells are mediated by transient complexes including hormonereceptor binding, signal transduction, correction of misfolded proteins by chaperones and allosteric enzymes which inhabit a brief interlude with co-partners [13]. A permanent complex is the highly stable ribosome, whereas regulatory pathways and signalling pathways are mediated by transient binding of protein-protein interactions. Such transactions are further subdivided into strong and weak binding. Heterotrimeric G protein is an example of a strong transient binding which is the result of an equilibrium shift between association/dissociation dependent on the activation of specific triggers in the protein pathway [13]. Both groups are separated on the basis of two characteristic features: time and stability. A permanent interaction is stable and will generally be irreversible. In contrast to transient protein interactions which are brief and where proteins will frequently associate and dissociate in the complex. Factors which trigger association/dissociation to occur, can be due to chemical modifications or conformational change in the protein, and co-localisation are all known to cause transient protein interactions [9-11]. A good example to illustrate the point is the complex formed by Heterotrimeric G protein bound to either GDP/GTP an example alluded to earlier. Complexes with transient oligomerization enable a protein to engage in multiple different interactions which can cause a change to the oligomerization state at any point in time. A weak transient complex requires minimal stimuli, such as a change in the concentration of pH or temperature, which can alter the state of the oligomer [9]. Weak associations have interfaces that are small and planar. On the other hand, a strong transient interaction can be triggered by a strong stimuli. An example is the GTP/GDP exchange or a phosphorylation event which can drastically alter the physiochemical and geometrical structure thus influencing a permanent complex to dissociate (Figure 8) [9,13].
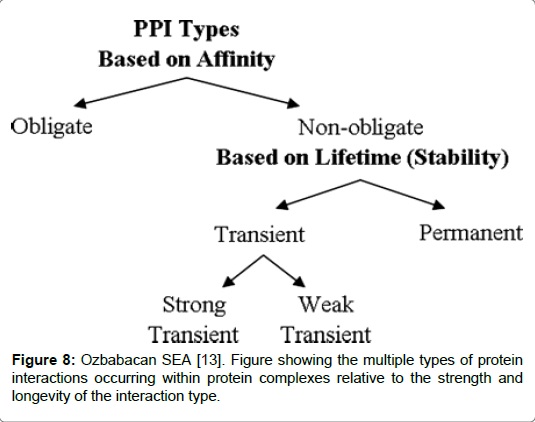
Figure 8: Ozbabacan SEA [13]. Figure showing the multiple types of protein interactions occurring within protein complexes relative to the strength and longevity of the interaction type.
Non-Scaffold Proteins
The development of non-scaffold proteins is a relatively new area in scientific research. The last five years has seen an increase in the use of non-scaffold proteins in both academia and in industry [18]. For more than a century antibodies have dominated the area of protein binding and have been used successfully in clinical application to treat a variety of diseases such as cancer, cardiac and infectious diseases [19,20]. Since the time Hybridoma technology was first introduced by Kohler and Milstein access to monoclonal antibodies (mAbs), as a tool in scientific research and in drug therapy, for the pharma industry became possible [18]. Its launch has resulted in the Food and Drug Administration (FDA) approving in excess of 20 antibody based pharmaceuticals for the treatment of disease. Moreover, with advances in DNA technology it has been possible to engineer antibodies in-vitro without using mouse model immunisation methods. Moreover, its use in research as well as in diagnostic and chromatography applications has been well documented [18]. Furthermore, non-Ig scaffolds have been used in the treatment as well as diagnosis of cancer and inflammatory disease.
In the last four years, a total of 20 different types of non-Ig scaffolds have been developed and these include: Adhirons, Alphabodies, Centyrins, Pronectins, Repebodies and Affirmers are just a few of the non-scaffold proteins currently in use. The single domain proteins are easy to manufacture and have a relatively quick turnover time and take approximately seven weeks to generate. This is due largely to the simple design and the absence of post- translational modifications that lack the presence of disulphide bonds. Non-Ig scaffolds have been used in monitoring post-translational modifications and for replacing antibodies in microscopy, flow cytometry and in western blot molecular techniques [20].
Antibodies in Protein Studies
The use of antibodies in molecular biology to tag proteins of interest (POI) in a cell is the most regularly used technique to target endogenous proteins in cell biology [21]. The technique called immunolabeling involves labelling a protein of interest (POI) with a primary antibody and binding a secondary antibody to the primary antibody in order to amplify the signal. The secondary antibody can often be conjugated to a small organic dye or a quantum dot (QD). Alternatively, primary antibodies can be directly conjugated to either a fluorophore or to a biotin molecule which can provide an alternative signal [21]. Quantum Dots (QDs) are nanocrystals and at discrete wavelengths have the ability to fluoresce concomitant to its size, QDs can provide a good quantum yield and have high coefficients, which are usually 10 to 100 times better than the small fluorophores and fluorescent proteins (FPs). Since coating QDs they have been successfully conjugated to antibodies and streptavidin thus making the biological application water soluble which can assist in eliminating quenching [21]. An area in which antibodies have been used with promising success is the field of drug therapeutics. Immunoglobulins (Igs) have been used as biological drugs, the reason behind its success lies in the short generation time against a specific target which can be an antigen or a hapten. Antibodies can be generated via the classical pathway or from a cloned or synthetic library. In the classical pathway a mouse or other animal is immunized with a specific antigen, the mouse produces antibodies against the injected antigen. It has been suggested that the specificity of an antibody for its target can be extremely high with an affinity in the low nanomolar or picomolar range which is far superior to most current drugs [19]. However, Phizicky et al. [22] argues that in analytical microarray applications antibodies lack specificity and quantification. He further states that a caveat with antibodies is cross reactivity with other proteins as well as to the protein/antigen of interest leading to unreliable results. Crystal structures of antibody Fab fragments complexed to antigen depict a similarity of antibody with antigen binding and that of protein-protein interactions. They reveal complementary binding between the interacting surfaces and with polar residues leading to the formation of hydrogen bonds [23]. An example of a crystal structure is the interaction between a Fab fragment complexed with an enzyme lysozyme as well as two Fab fragments interacting with the influenza virus surface protein neuraminidase [23]. The use of antibodies does have its limitations which centre on its large size, as well as the complexity of generating four individual protein chains, that require glycosylation of the heavy chains and the presence of a disulphide bond in each of the several Ig domains [18,19]. A widely used antibody type is the IgG molecule, a bivalent and multidomain protein that relies on a complex glycosylation process and disulphide bonds.
One of the main problems with the protein molecule is that it is unstable at high temperatures and also expensive to manufacture. The high cost are inextricably linked to the complexity of antibodies. Each antibody is made up of two heavy chains and two light chains that are post translationally glycosylated, and as a result generating antibodies in microbial cells can be a challenge. Antibodies are not easy to synthesise and require cells to have folding chaperons and specific glycosyl transferases that are present only in mammalian cells. The time taken to generate antibodies can be anything between six months to a year which can cause significant delays particularly in research and can put back results data causing significant delays (Figure 9) [24].
Figure 9: An IgG molecule comprised of two heavy chains and two light chains. Heavy chains are made up of three constant domains (blue) and one variable domain (VL light green) Helma J [25].
Introducing Non-Protein Scaffolds
The generation of protein scaffolds utilises the molecular recognition and specificity of antibodies, with an even better design and with improved characteristics, which include a small size, improved stability at higher temperatures and an absence of disulphide bonds. Bacterial expression systems are used to generate the protein scaffold which can yield vast quantities. The favourable characteristics and better design has seen a few protein scaffolds enter clinical trials. The prevailing view is to design a small subset of protein scaffolds that can be used against a diverse number of targets in varied settings. The idea is to develop protein scaffolds that can be used as a drug therapy or for use as diagnostics. The main scaffolds are: Adnectins, Affibodies, Anticalins, DARPins are a few going forward as potential candidates for a new class of protein drugs [19]. In vivo imaging using SPECT (single-photon emission computed tomography) or PET (positron emission tomography) have shown that the small size of non-Ig scaffolds comes with some advantages. The small size allows better tissue penetration and a faster blood clearance. Amongst its uses non- Ig scaffolds include the use of DARPins which are targeted against extracellular signal regulated kinase 2 (ERK2) in both phosphorylated and non-phosphorylated states [20].
Adhirons
Adhirons were developed as alternatives to antibodies [25] and were designed by researchers at Leeds University in 2012. The strength and suitability of non-scaffold proteins lies in the design and structure of adhirons, its characteristic small size means it can penetrate tissues with a high degree of efficiency. The extremely robust scaffold makes it a paragon for protein scaffolds which can be produced efficiently and in large quantities in bacterial expression systems [25]. In determining the best possible structure for a protein scaffold that could potentially be used in therapeutics and in research a number of factors were considered by the team at Leeds University. The study looked into the design of Adhirons and assessed the possible caveats with using adhirons as therapeutics. In designing the construct thermostability was considered a factor furthermore, a correlation was also deduced between the stability of a protein and its thermostability. It is thought that stable scaffolds result in long term storage which can also be stored at ambient temperatures. This does bring potential benefits for heat purification and provides options for storage of reagents as well as for drug administration. The study further suggests that by inserting the loops between the β-sheets can cause the construct to become less stable however by choosing a stable scaffold can circumvent the structural weakness and can still be used effectively for library generation. A second consideration is that small molecules can have a short half-life which can reduce its effectiveness as a therapeutic model. However, the problem can be circumvented by binding the adhirons to large size proteins such as albumin or by using PEGylation or PASylation techniques [26]. The structure of andhirons commercially known by the name of Affirmer [25] is based on a sequence which is from a cysteine protease inhibitor called phytocystatin. The non-antibody scaffold demonstrates high thermal stability with a temperature (Tm) of 101˚C and shows good levels of expression in Escherichia coli. Its crystal structure has also been determined to a resolution of 1.75 Å which reveals a compact cystatin-like fold [26]. The molecule is classed as a novel antibody mimetic based on a protease inhibitor. It comes from a family of cystatins known for their highly conserved fold and which are made up of a central α-helix wrapped around by four antiparallel β-sheets. The scaffold has a truncated N-terminal and two inhibitory loops. One of the loops contains the sequence QXVXG which forms the active site. The loops are made from a set of nine random residues which form the variable region and are positioned between the β-sheets. The randomised amino acids in each loop replace the inhibitory sequences within the Gln Val Ala Gly and Pro Trp and Glu loops of the consensus sequence of phytocystatin [26]. The insertion of the two variable regions in the adhiron molecule forms the basis of the phage-display library which is made up of 1.3 x 1010 clones. The phage library was tested against the yeast Small Ubiquitin-like Modifier (SUMO). It was shown that the variable region 1 contained sequences which were homologous to SUMO interactive motif (V/I-XV/ I-V/I). Further characterisation of adhirons resulted in the selections of four adhirons that demonstrated no cross reactivity to human SUMO protein isoforms however, they did show high specificity and low nano molar affinity to the yeast SUMO [26]. Cystatins function as competitive inhibitors and are found in nearly every form of life. They act by binding to the active site of cysteine proteases as pseudosubstrates and render them unable to cleave peptide bonds. It has been suggested from studies, that the protein found in plants, contains many important properties which are thought to confer an advantage to both plants and humans. In host plants defence systems are expressed in response to stress, resulting from plant wounding or in response to pest infestation. It is thought that they cause a deficiency in proteins which can reduce pest survival and slow down development. The proteins have been used in the control of fungal and viral pathogens by targeting virus replication. However, the exact mechanism by which fungal pathogens are controlled has yet to be elucidated [27]. Cystatins have the potential for future applications such as in the protection of genetically modified transgenic plants as well as in disease management. Its use in plant based recombinant protein applications has provided promising results. Protein engineering techniques are being used on plant protein cystatins to improve specificity and potency. By engineering new amino acid sequences in the conserved protein regions or in the variable amino acid sites increased potency. Studies have shown that by mutating the N-terminus has the potential of improving protein flexibility by disrupting the hydrophobic cystatin core. The aim to increase flexibility in this region will decrease stearic hindrance by increasing the binding of the two inhibitory hairpin loops at the catalytic site (Figure 10) [27].
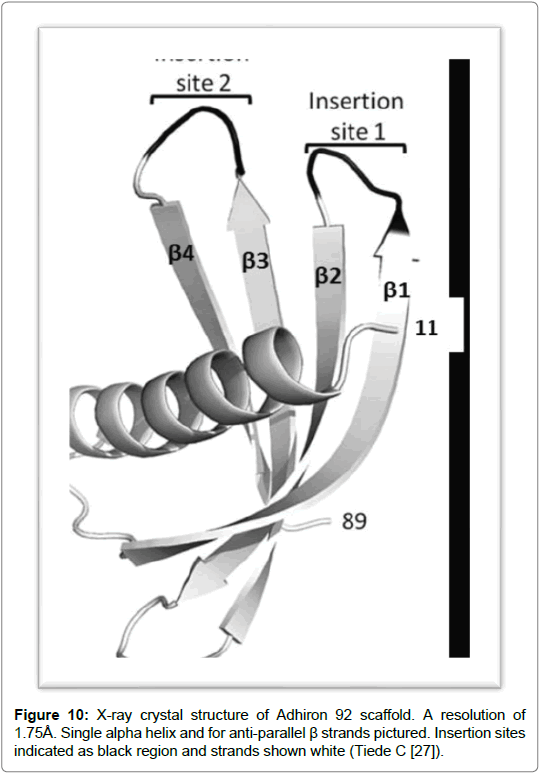
Figure 10: X-ray crystal structure of Adhiron 92 scaffold. A resolution of 1.75Å. Single alpha helix and for anti-parallel β strands pictured. Insertion sites indicated as black region and strands shown white (Tiede C [27]).
Research into cystatin has led to the development of plant cystatin based novel non-antibody scaffold proteins called Adhirons. Researchers at Leeds University believe that adhirons will one day replace antibodies in scientific research and in diagnostics due to its excellent characteristics. Its small size and monomeric structure enabling increased solubility and a high degree of stability. The adhiron molecule lack disulphide bonds and glycosylation sites, both of which are required by antibodies for stability. Unlike antibodies, adhirons can be expressed easily in bacterial systems thus making Adhirons serious contenders for use in biological applications. Magnetic Interacting Adhirons (MIA) were used in identifying the interaction of specific proteins with the (100) faces of cubic magnetite nanoparticles. The study showed that basic amino acids lysine had the lowest adsorption energy when interacting with the nanoparticles. Moreover, that the lysine residue can direct the cubic shape of the nanoparticle [25]. MIA performed better in making cubic nanoparticles at room temperature. This contrasted to the high temperatures and harsh conditions used previously to form cubic nanoparticles. Studies have also found the identity of binders against a number of targets which include the fibroblast growth factor (FGF1) against platelet endothelial cell adhesion molecule (PECAM-1) [26].
DARPins
A type of non-Ig scaffold gaining notoriety is DARPin. It has been suggested that in the future these protein scaffolds may replace antibodies either partly or completely [28-30]. Limitations in the design and the use of monoclonal antibodies in research and diagnostics has led to the search for finding better alternatives. The aim has been to find alternatives which can be produced easily, are cost effective and have a shorter turnover time [31,32]. DARPins are a class of protein scaffolds that fall under the group called repeat proteins [28]. They are from natural Ankyrin repeat proteins which are the most abundant binding protein found in the human genome and can bind to different proteins resulting in different biological reactions [31,32]. The structure is made up of two to four randomised repeats that are genetically fused. The repeats are flanked by N-and C- capping repeats which facilitate folding and prevent protein aggregation. DARPins are composed of 33 amino acids made of both α-helices and β-sheets. The scaffolds are approximately 10% of the size of an antibody and have a mass of anywhere between 14 to 21 kDa [32]. The scaffolds possess structural characteristics that confer an advantage over antibodies. The small size allows rapid tissue penetration where a larger sized antibodies can find it challenging to penetrate the barrier of membrane proteins. Another advantage is that they can be rapidly cleared from the bloodstream which limits the risk of toxicity. Therefore DARPins can be used in therapeutics such as in drug therapy [33]. DARPins have been used successfully in crystallography studies. The binding of DARPin with Maltose Binding Protein (MBP) from bacteria Escherichia coli was the first proof-of-principle example involving the crystallisation and selection of a DARPin in complex with a target protein [30]. Moreover, in the same study (2008) the structure was solved at 23 Å resolutions. Determining the structure was achieved by using a previous known example of an unselected DARPin that had been solved (Figure 11) [30].
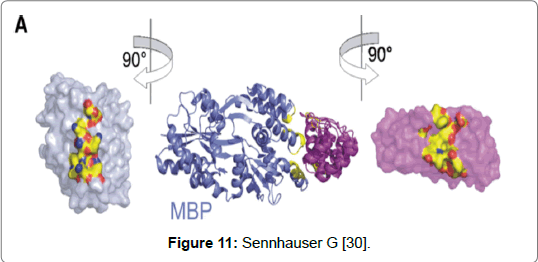
Figure 11: Sennhauser G [30].
An important consideration for using protein scaffolds is that the technique can produce a crystalized structure of a target easily and efficiently. Two technologies used in co- crystallisation studies involving DARPins are Ribosome Display (RD) and Phage Display (PD) both have large library selection technologies. The ribosomes display an in-vitro technique that forms non-covalent ternary complexes from ribosomes, mRNA and nascent protein chains. These complexes are then used as a test for binding to a target protein. It was possible to determine the structural complexation between the selected DARPin and MBP due to the relative size of the DARPin (~ 18kDa) to the MBP (~43kDa). The structure determination showed that upon complexation that three helices at one side of the elongated MBP, were able to bind to off7 and that a positive surface patch which was made up of four lysines and which formed 60% of the buried surface area was in close complexation with off7. Furthermore, three framework mutations including H125Y were found to be in off7. The DARPin’s interface is characterised by seven aromatic residues of which four are tyrosine residues and upon complexation represent 70% of the buried surface area. The use of DARPin’s in drug therapy has been studied and with good results. DARPin’s are single domain proteins with an absent effector function however, effector functions can be added to DARPins in number of different ways. The advantage of lacking a cysteine molecule does mean that a site specific thiol group can be added which has no effect on the binding interaction. The moiety can either be a cytotoxine or a radioactive isotope. The small size of DARPins does confer advantages over antibodies in that DARPin’s are able to penetrate tissues more easily than antibodies. Secondly the specificity with which DARPins can target disease tissues and its relative short serum half-life mean that it can accumulate in disease tissue. However, simultaneously unbound molecules can be removed via the kidneys. Due to the obvious benefits the use of DARPins in the treatment of cancer holds promise [31].
Repebodies
The application of repebodies in protein interactions is a new phenomenon which has only recently been used. To date, antibodies have dominated the field of proteomic studies and have been the primary choice as molecular binders tracking protein interactions and for in vivo protein localisation studies in cells. Antibodies have a number of drawbacks which negate their use in proteomics studies. The large size of antibodies means they can be poor at penetrating target tissues and have low expression in cells creating significant problems. Due to the issues of antibodies it can cause limitations in the analysis of protein-protein interactions. For this reason small protein scaffolds termed repebodies were developed. A study by Kim et al. was designed to demonstrate the utility of repebodies in protein-interaction studies. It was shown by tracking a rapamycin-mediated interaction that involved two proteins the FKDP 12–rapamycin binding (FRB) domain and FK506-binding protein (FKBP) in cells. A repebody was designed using phage display and a high affinity red fluorescent protein was fused to a green fluorescent protein to target and bind a protein in a mammalian cell bound to a red fluorescent protein. Repebody B1 had specificity for mOrange and was designed from five rounds of bio-panning. By introducing two mutations LRRV2 and LRRV into two different modules a repebody specific for red fluorescent protein was developed. The Kd was determined at 31.9 nM and 40.0 nM for mOrange and mCherry respectively and which exhibited little or no cross talk with green or yellow fluorescent protein (Table 3).
| Clone Name | Target | N | ∆H(kcal mol | ∆S (cal mol-1 deg-1 ) | Kₒ (nM) |
|---|---|---|---|---|---|
| Repebody-B1 | mOrange | 0.92 ± 0.01 | -30.2 | 31.9 ± 11.7 | |
| -19.33 ± 0.29 | -15.6 | 40.0 ± 9.1 | |||
| mCherry | 0.96 ± 0.01 | ||||
| -14.78 ± 0.16 |
Table 3: Data showing dissociation constant and binding energetics of Repebody-B1.
The results of the study determined a number of key indicators that assessed if the repebody -B1 was successful in binding to its target protein. To test the binding of repebody-B1 to its target, Size Exclusion Chromatography (SEC) was used. It was shown that the two proteins could form a complexation at a lower elution volume and were able to interact in a reducing environment. Therefore, demonstrating that the repebody-B1 can bind and recognise its target protein in the reducing intracellular environment of mammalian cells. The second determinant was co- localisation and which was assessed by fusing EGFP to repebody-B1 as a reporter protein expressed in Hela cells. Confocal microscopy results showed high levels of expression and distribution of the fusion protein in the nucleus and across the cytosol in mammalian cells. Furthermore, co-localisation or complexation of EGFP-repebody-B1 to its target mCherry in mammalian cells was demonstrated using End Binding Protein (EBP), a core component found in microtubule-plus end tracking protein network. EBP was fused to mCherry and EGFP repebody-B1 was co-expressed in Hela cells, the analysis of complexation was demonstrated using confocal microscopy. A comet-like pattern at the microtubule end with green and red fluorescent protein merging appeared to show complexation. The results suggested that EGFP repebody-B1 complex recognised mCherry in the subcellular compartment (Figure 12). The study examined EGFP-repebody-B1 and its usefulness in protein-interactions studies in living cells. To assess whether FKBP (FK 506-binding protein) and FRB (FKBP12- rapamycin binding) domain could interact was demonstrated using mCherry fused to FKBP and the FRB domain fused to myristolylation signal motif (Myr) of Lyn and by anchoring the FRB domain to the cytoplasmic membrane. In the presence of rapamycin the FRB domain recruited FKBP-mCherry to the plasma membrane suggesting that in the absence of rapamycin the cytoplasm expressed both FKBP mCherry and EGFP-repebody-B1 equally, however after adding rapamycin FKBP mCherry and EGFP-repebody-B1 colocalised to the plasma membrane. These results demonstrate the utility of using repebody-B1 in protein interaction studies and in targeting specific proteins in mammalian cells (Figure 13).
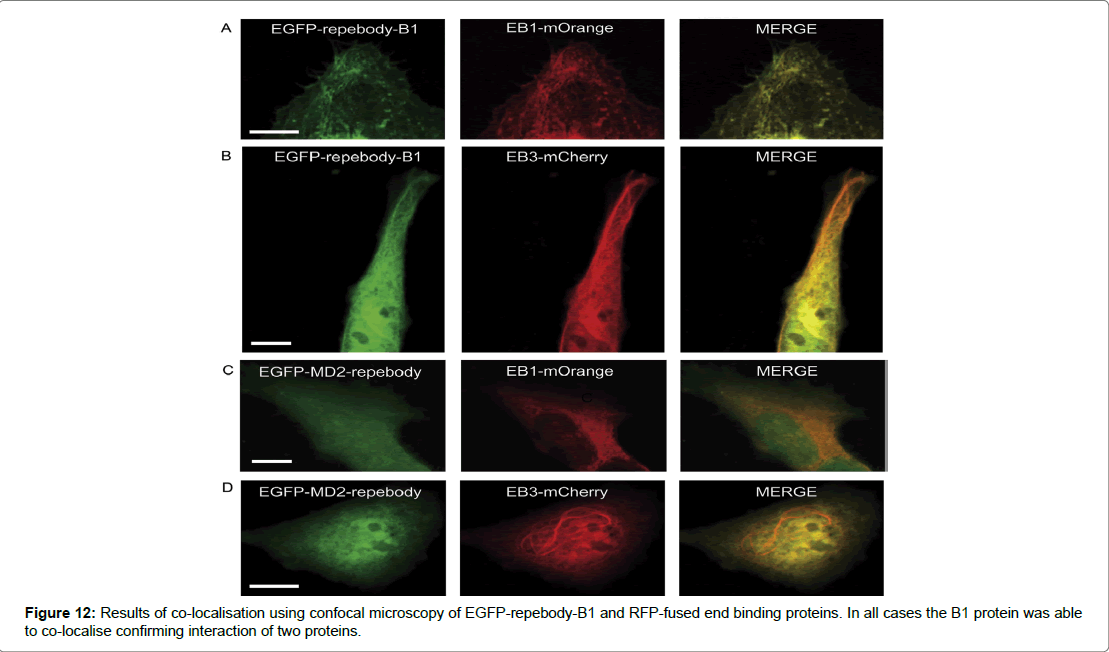
Figure 12: Results of co-localisation using confocal microscopy of EGFP-repebody-B1 and RFP-fused end binding proteins. In all cases the B1 protein was able to co-localise confirming interaction of two proteins.
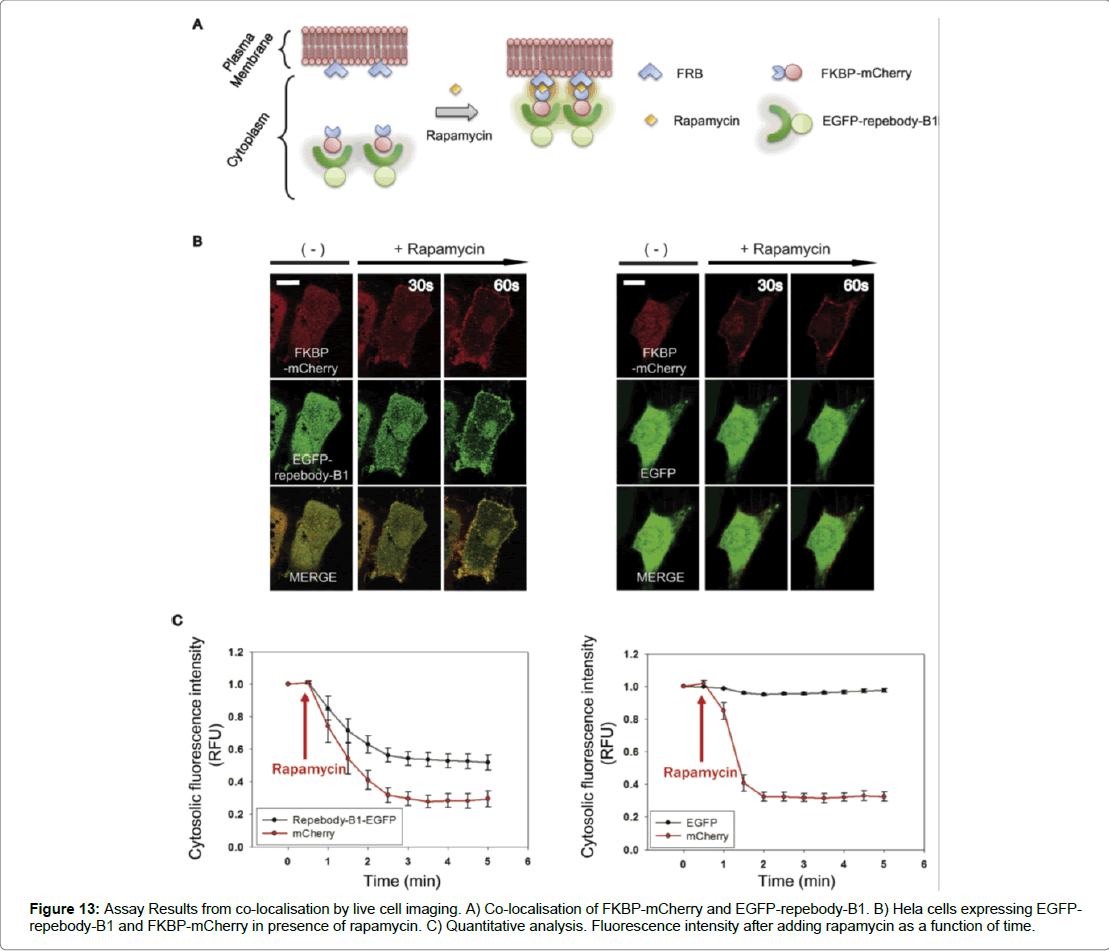
Figure 13: Assay Results from co-localisation by live cell imaging. A) Co-localisation of FKBP-mCherry and EGFP-repebody-B1. B) Hela cells expressing EGFPrepebody- B1 and FKBP-mCherry in presence of rapamycin. C) Quantitative analysis. Fluorescence intensity after adding rapamycin as a function of time.
The structure of repebodies is derived from consensus design leucine-rich repeat (LRR) molecules. Favourable properties are highly stable at a number of different pH ranges and temperatures and express well in Escherichia coli [33]. Repebodies are used as therapeutics and show potential for use in the treatment of AMD (Age related macular degeneration) a leading cause of blindness and vision loss in the over sixties. Other candidates for therapy have been diabetic retinopathy and metastatic cancers. Repebodies have shown efficacy in inhibiting the VEGF pathway and in blocking cellular processes such as proliferation and migration. By developing a prototype repebody which has an antiangiogenic agent named anti-human VEGF repebody selected via phage display, the rC2 repebody type displayed the highest affinity for human VEGF and was shown (Figure 14) [34].
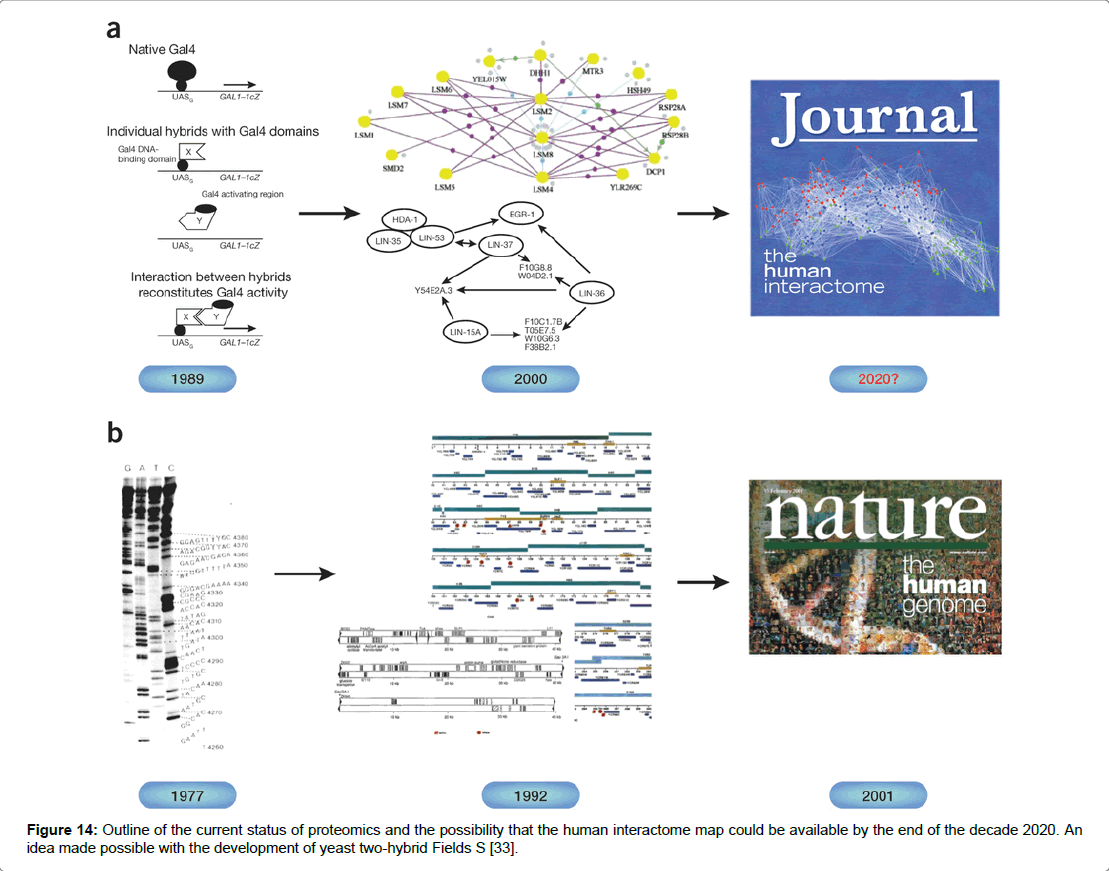
Figure 14: Outline of the current status of proteomics and the possibility that the human interactome map could be available by the end of the decade 2020. An idea made possible with the development of yeast two-hybrid Fields S [33].
Yeast Two Hybrid
A high throughput method used by researchers in protein–protein studies is the yeast-based genetic assay yeast-two hybrid (Y2H) for the detection of in-vivo protein interactions [35]. The in vivo assay was originally developed by yeast geneticist Fields in 1989 [36]. The technique utilised the interaction of a pair of proteins enabling the transcription activation domain (TAD) and the DNA binding domain (DBD) to come into close proximity resulting in the activation of an adjacent reporter gene [37]. The technique performed in yeast cells involved the construction of a hybrid gene which encodes a protein X fused to DBD and a protein Y fused to TAD. Transcription is activated when protein X fused to TAD binds to protein Y which fused to DBD. Where the target protein Y is not fused to DBD a protein interaction does not activate transcription [34]. The central concept is that the DNA binding domain and the Activation domain act independently. Acting alone the domains cannot activate transcription led to other possibilities that any pair of proteins fused to DBD and TAD could reform the transcriptional activator via a two protein interaction [38]. The bait and prey proteins are expressed simultaneously in the nucleus under the control of a transcription factor GAL4 activating transcription of genes. Protein X and protein Y are used to bring DBD and TAD together and to restore the activity of the transcriptional activator [39]. It has been suggested that the two hybrid system was designed to debunk the biological circuitry apparent in most diseases such as cancer and heart disease (Figure 15) [34,39]. The technique widely employed in assay studies has received a fair amount of negative criticism which centre on the efficacy of the data produced and its reliability. The main caveat is the high numbers of false positives which can occur [34,38]. However, it has been suggested that false positives are not due to flaws in the design of the tool but rather in the methodology used by the researcher. The same study by Vidal and Fields (2014) further points out that readout depends on cells being able to grow and that the growth of artefacts can be unrelated to the formation of the transcription factor. Occasionally mutations or rearrangement can cause a hybrid of DNA binding domain to self-activate without reconstitution of the transcription factor. It has been suggested that the interactome will be fully mapped by 2020 and that the yeast-two hybrid technique will be used in that effort [34].
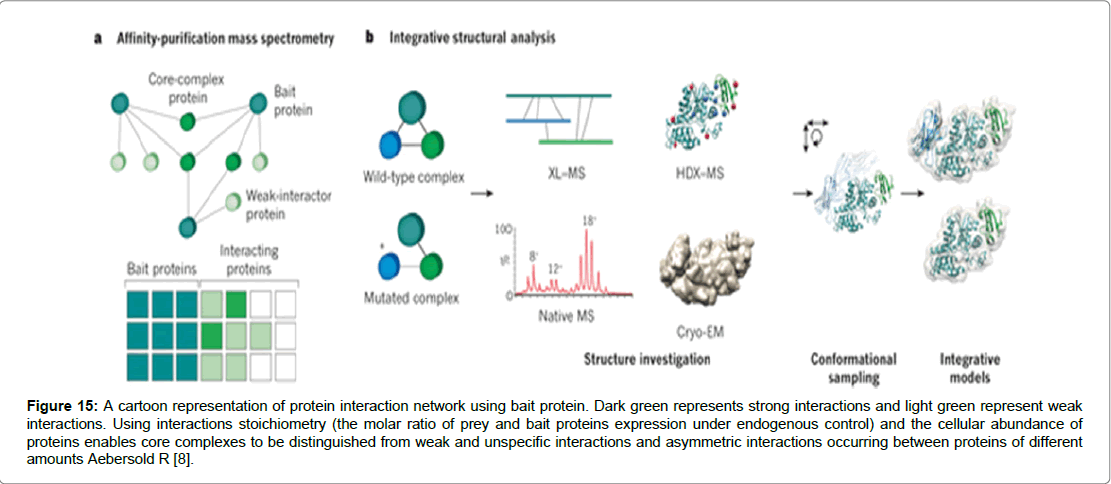
Figure 15: A cartoon representation of protein interaction network using bait protein. Dark green represents strong interactions and light green represent weak interactions. Using interactions stoichiometry (the molar ratio of prey and bait proteins expression under endogenous control) and the cellular abundance of proteins enables core complexes to be distinguished from weak and unspecific interactions and asymmetric interactions occurring between proteins of different amounts Aebersold R [8].
Mass Spectrometry in Protein Interaction
The potential of Mass spectrometry (MS) a highly sensitive technique which bridges the gap that exists in the analysis of proteins in cells and tissues. The study of de-novo proteins is widely seen as a challenging area of proteomics due in part to the low levels of proteins found in cells and tissues [39]. Therefore the necessity to design tools and techniques that are highly specific is central to biological studies in protein interactions. MS is viewed by many as meeting the challenge for two reasons. It has widely available databases on gene sequences and second it developed protein ionization [39]. Mass spectrometry ionization technique that won the Nobel Prize in Chemistry in 2002 and is at the heart of mass spectrometry [40]. Essentially the technique comprises three essential aspects: an ion source, a mass analyser and an ion detector. The first stage is conversion of molecules to a gas-phase ions. Molecules must transfer from solution as solid phase into gas phase as ions. A mass analyser can separate ions on the basis of mass to charge ratio. However, it is possible to manipulate and direct ions to a detector in an m/z dependent manner via electric or magnetic fields. As the ions strike a detector an image is created which records the number of ions and ascribes an m/z value [39-41]. Two techniques which have been used abundantly to volatise and ionise protein and peptides in readiness for analysis by MS are the widely employed Electrospray Ionization (ESI) and Matrix-assisted laser desorption/ ionisation (MALDI) [39]. MS technology has been used collaboratively with pull down assays that involve bait and prey proteins in a binding complex. The strategy known as affinity-purification mass spectrometry (AP-MS) employs a pull down assay of the bait protein bound to its target protein proceeded by mass spectrometric analysis. In order to be able to distinguish between a bait protein binding specifically to a partner protein and contaminants causing non-specific binding, essential controls are put in place that allow quantitative comparison of samples (Aebersold and Mann, 2015). The bait and prey technique enables binding stoichiometries to be classified into three quantitative dimensions of stable, regulatory or transient interactions (Figure 16) [42,43]. The complexity that comes with post-translational modifications and the relative differences in protein abundance. As well as the regulation of protein modifications, governed by time and context, mean that current MS tools require upgrading to meet the demands of proteomic studies [42]. It has been suggested that a faster scan speed would increase the amount of sampling ions resulting in more tandem mass spectra acquired per unit time. Secondly by using larger sample sizes would increase the range as lower abundance ions could also be detected. Thirdly by improving sensitivity and mass accuracy could lead to achieving higher confidence levels in identifying peptides and the resulting interactions [42,44].
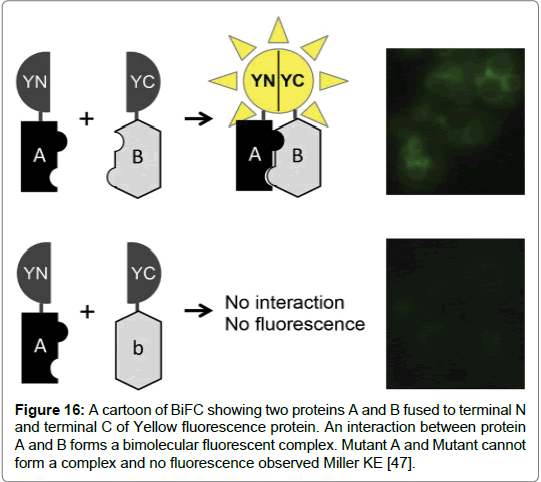
Figure 16: A cartoon of BiFC showing two proteins A and B fused to terminal N and terminal C of Yellow fluorescence protein. An interaction between protein A and B forms a bimolecular fluorescent complex. Mutant A and Mutant cannot form a complex and no fluorescence observed Miller KE [47].
Bioluminescence Resonance Energy Transfer (BRET)/ Fluorescence Resonance Energy Transfer (FRET)/ Bioluminescence Resonance Energy Transfer (BIFC)
The central pillar connecting the three biological fluorescence and luminescence based approaches to protein-protein interactions studies is the ability to characterise the spatiotemporal aspect of proteinprotein interactions (PPIs) [45]. All three techniques are regarded as powerful approaches by providing the spatiotemporal data of PPIs in live cells. The advantage has been that it has provided native proteinprotein interactions (PPI) without using mechanical or detergent based cell lysis methods [45]. The development of non-invasive fluorescencebased methods of FRET, BRET and BIFC are tools that allow the visualisation of the behaviour of proteins in their native environment [46]. The fundamental facets of BRET is the transfer of a non-radiative energy between a donor and acceptor. In contrast FRET uses two fluorophores, a donor and an acceptor fluorophore. The donor absorbs exogenous excitation and transfers the energy to the acceptor fluorophore [46,47]. The transfer occurs via dipole-dipole coupling with one important aspect, the transfer distances occur over 10 Å and 100 Å. A second determinant is that the fluorescence spectrum of the donor and the absorbance spectrum of the acceptor sufficiently overlap and both the quantum yield of donor and the absorption coefficient of acceptor are adequately high [46]. The BRET technique involves the transfer of energy derived from an enzyme, which during catalysis and via oxidation of its substrate luciferase, transfers the energy to the acceptor molecule. It is considered similar to FRET, in that both techniques do not depend on an external light source. Moreover, that both BRET and FRET function when the emission spectrum of donor and excitation spectrum of acceptor have sufficiently overlapped [47]. The success of BRET in protein interaction studies relies on the distance between donor and acceptor which is why it has been used extensively to image protein-protein complexation. The success of FRET based techniques enables proteins fused to GFP to target proteins with signal sequences to specific compartments in the cells. Thus it enables the in-vivo visualization of specific protein interactions as they occur in cells and in their native environments. However, the technique has revealed caveats such as photobleaching of the donor fluorophore as well as auto fluorescence of cell and tissues and damage to cell/tissues by excitation light. In addition, further issues can cause stimulation of tissue, particularly in the case of the retina, which is a highly photoresponsive tissue. Further, it is known to cause excitation of the acceptor fluorophore that is unrelated to resonance transfer. However, the limitations seen in FRET are not the same for BRET technique which does not have a requirement for excitation light to function. In BRET the donor fluorophore used is luciferase the bioluminescence from luciferase when substrate is present can cause excitation of the acceptor fluorophore via the same resonance energy transfer route as FRET [46].
Bimolecular Fluorescence Complementation (BiFC) is a biological tool increasingly used in the study of protein-protein interactions. The in-vivo tool enables high through-put screening of protein-protein interactions as well as the visualisation and analysis of drug interactions that regulate protein- protein interactions (PPI) [48].
The essence of BiFC is the in vivo reconstitution of a fluorescent protein has proven quite successful in revealing protein interactions and insights into protein functions. BiFC mimics the method used by Protein Fragment Complementation Assays (PCAs) where a reporter protein is first truncated before being fused to two proteins of interest (POI). The reporter protein can either be an enzyme or a fluorescent protein. In BiFC the reporter protein used is a green fluorescent protein (GFP). GFP is first split into two parts. Two target proteins are fused to the N- terminal and C- terminal of the GFP protein expressed in cells. When the two target proteins interact the GFP protein parts come together forming a GFP complex. Using Flow cytometry and fluorescence microscopy the signal given by BiFC technique can be visualised (Table 4) [48].
| Method | Application | Disadvantage/Advantage | Therapeutic uses |
|---|---|---|---|
| Antibodies | Targets endogenous proteins in cell | Large size | Biological drugs either as polyclonal or monoclonal antibodies |
| Can be labelled with primary or secondary antibody | High cost and complicated to generate due to glycosylation of heavy chains | Extensive use in research as in western blot, flow cytometry, Immunohistochemistry (IHC), Enzyme-linked immunosorbent assay (ELISA) | |
| Secondary antibodies conjugated to biotin or fluorophore | Un stable at high temperatures | Biomarkers for diagnostics | |
| Can take between six months to a year to generate | Used in Oncology and inflammatory and viral diseases | ||
| Not easy to synthesise require that mammalian cells to have chaperones e.g. glycosyl transferases | 500 k different antibodies available | ||
| Difficult express in bacterial system | |||
| Adhirons | Expressed in Escherichia coli | Advantages; small size | Diagnostics and generation of chemical agents |
| Scaffold contains phage display library of different proteins | Easily penetrates tissue where antibodies too large | Used academic research for studying protein- protein interactions studies | |
| Library contains 1.3 X 1010 clones |
Disadvantages: Short life span | Cell signalling pathways | |
| Not ideal for use in therapy | Used in different biological applications: biosensors, ELISAs, Cell imaging, pull-down assays, affinity histochemistry, in-vivo Imaging | ||
| Can accumulate in tissue due to small size | |||
| DARPins | Made from repeat proteins called Ankyrin proteins | Advantage: abundant proteins | Drug therapy |
| Can be used in different biological reactions | Crystallography studies | ||
| Generated with 2 to 4 randomised repeats | Small size, rapid tissue penetration | Tumour targeting | |
| Made of 33 amino acids of both α and β- sheets | Easily cleared from blood | Used in biological applications: IHC, HER2 gene amplification status for detecting optimum therapy in breast cancer treatment | |
| 10% of size of antibody | Low levels of toxicity | In diagnostic pathology | |
| Highly stable proteins due to repeat protein design | |||
| Repebodies | Made up of leucine-rich repeats (LRR) module | Advantage: Due to expression in Escherichia coli therefore relatively inexpensive to make | Protein binding and localisation |
| Generated via a polypeptide by fusing N- terminus of internalin protein | Can be generated in large amounts | Treatment in age related macular degeneration | |
| Expressed in Escherichia coli | High physical and chemical stability | Diabetic Retinopathy | |
| Novel protein and free from existing patents | Metastatic cancers | ||
| Have high affinity to VEGF | |||
| Good disease biomarker | |||
| Yeast 2 hybrid (Y2H) | Involves the interaction of pair of proteins each pair binds either to TAD (Transactivation domain) or DBD (DNA binding domain) | Disadvantage: High numbers of false positive results | Used in cancer and heart disease studies |
| Pair of proteins must bind to each other for transcription to be activated | Mutations or DNA rearrangement can cause hybrid of DNA binding domain to self-activate without first binding to partner protein | Protein interactions studies with some success | |
| Mass spectrometry | Based on ionisation technique and uses ion source, mass analyser and ion detector | Protein interactions studies and determining identity of proteins | |
| Can be used with Electrospray Ionization (ESI) and Matrix assisted laser desorption (MALDI) | Advantages: High resolution and high mass accuracy | Used in biological application such as pull- down assays | |
| Different types of Mass spectrometry applications: Orbitrap Fourier transform mass spectrometry and Time-of –flight mass spectrometry | Drug testing/ drug screening | ||
| Forensic toxicology | |||
| Clinical toxicology | |||
| Used in structural studies | |||
| FRET/BRET | Luminescence based and fluorescence based methods | Advantage; Detection of native proteins without the need to use cell lysis methods | |
| Involves the transfer of non-radiative energy between a donor and acceptor (BRET) In FRET uses two fluorophores, an acceptor and donor a transfer of energy occurs between the two fluorophores |
Disadvantages: photo bleaching and auto fluorescence of cell and tissue Can cause damage to tissues and cells due to excitation energy of light |
||
| BIFC (bimolecular fluorescence complementation) | Uses a truncated reporter protein and fused to protein of interest. The reporter protein is an enzyme or a fluorescent protein | Advantages: fluorescent technique does not require that cells are first pre-treated with cell lysis or fixation | High through-put screening of protein-protein interactions |
| High sensitivity and minimal background noise | Used in drug interactions | ||
| Require the use of good negative controls | Protein function studies |
Table 4: A list of current biological tools used in protein interaction studies.
The technique has advantages. As it is a fluorescent technique the application does not require cells to be pre-treated with cell lysis or cell fixation. Therefore, visualisation of PPI’s is possible with minimal cell disturbance. Secondly it is a highly sensitive method that gives minimum background noise as the fragments of YFP require two proteins to complex before fluorescence can occur. The main issue with the application of BiFC is the spontaneous folding of the fragments of fluorescent protein. Moreover, the intensity of fluorescence for the recombined fragments should be equal to the fluorescence of an intact protein in order to discern background noise. Good negative controls are used in order to test positive interactions however, finding negative controls to use presents challenges (Figure 16) [48].
DNA Paint under the Spotlight
An important tool in microscopy is optical microscopy for studying living cells and organisms. However, it has caveats particularly in regards to spatial resolution of the image caused by the limitations arising from the diffraction of light [49]. The optical microscope allowing visualisation of cellular protein distribution is only visible for a certain time. Recent developments in biological tools such as “super-resolution” far field optical microscopy (nanoscopy) challenges current limitations in spatial resolution. Techniques such as stimulated emission depletion (STED), ground state depletion (GSD), reversible saturated optical (fluorescence) transitions (RESOLFT) and photoactivation localisation microscopy (PALM) are just few of the nanoscopy techniques available Hell et al. STED, RESOLFT, PALM/ STORM push optical techniques to an optical resolution of nanometer scale. However it has been suggested that even though the optical resolution is high the techniques offer a degree of technical complexity and therefore multiplexing a distinct number of targets can be equally technically challenging [49,50].
Point accumulation for imaging in nanoscale topography (PAINT) is seen as an alternative to the highly involved methods. It uses diffusing fluorescent molecules that transiently interact with the sample. The method is described as easy to operate and requiring no special conditions or equipment to achieve photoswitching [50]. It has been observed that a key limitation with PAINT is the interaction of dyes with sample via electrostatic coupling or hydrophobic interactions. The caveat with the technique reduces the availability of the dye limiting simultaneous imaging of biomolecules of interest [50].
A further adaption to PAINT is termed universal PAINT which addresses the issues by simultaneously and continuously labelling specific biomolecules with fluorescent ligands such as antibodies. Although the adaption facilitates specific dye sample interactions it fails to identify specific interactions with programmable kinetics [51].
One such method termed DNA-PAINT can achieve spatial resolution of approximately 10-nm in vitro and on DNA structures. A second technique termed Exchange- PAINT enables the imaging of multiple targets by using a single dye and laser source (Figure 17) [52-70].
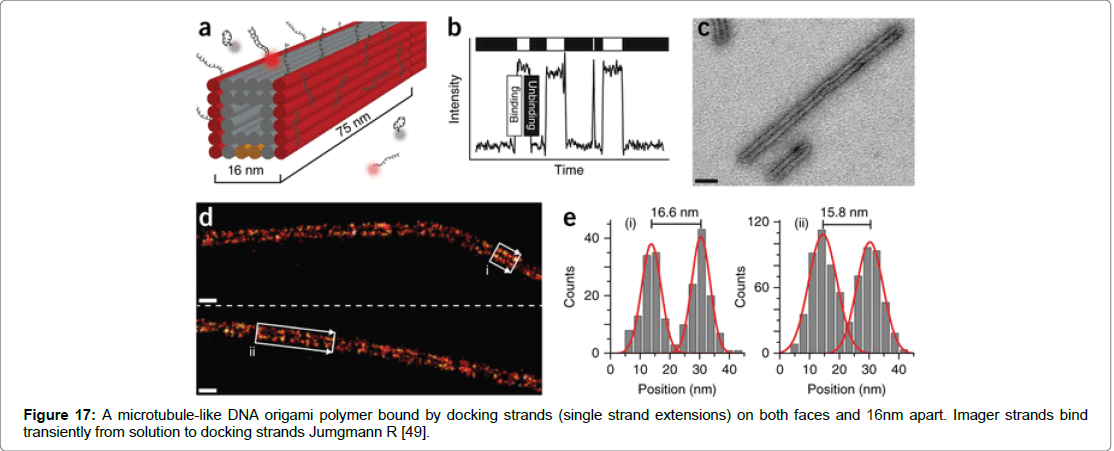
Figure 17: A microtubule-like DNA origami polymer bound by docking strands (single strand extensions) on both faces and 16nm apart. Imager strands bind transiently from solution to docking strands Jumgmann R [49].
DNA-PAINT achieves both high specificity and high number of usable fluorophores the technique involves stochastic switching of on and off states using fluorescence. The on/off switch is achieved via fluorescently labelled oligonucleotides called imager strands that bind to complementary docking strands of DNA nanostructures through repetitive and transient binding [71-85]. Spatial resolution is achieved within 25 nm and the technique has been used for multicolour sub diffraction.
To conclude the enormity of mapping the whole of the protein interactome will be a great achievement and technological challenge for science. This work was an attempt to address the fundamental aspects of protein interactions and the challenges of current tools employed in protein interaction studies. It was not the intention to cover the whole spectrum of technologies currently used due to the enormity of the field. The purpose was to explore and discuss the important factors and considerations in studying the interactome and to shine a torch on this very important gap in science. The study of proteomics presents the opportunity of combining techniques which is promising. The potential for mapping the signalling cascade of MAPK pathway can be achieved, it would open the way for mapping the interactome of more diseases and facilitate the development of new and innovative drugs for fatal diseases such as cancer, heart disease and pathogenic bacteria as well as viruses. The task will be costly, financially and in terms of time, both are valuable commodities. Scientist have suggested that by mapping the interactome following the human genome project, will be the next big scientific endeavour. However, there is no guarantee that disease causing proteins can be inhibited following mapping the whole of the protein interactome.
Research into protein interactions has shown that protein-protein interactions govern and regulate all biological processes in cells. Time will reveal whether science can achieve this important milestone. Sequencing the genome was once an idea that that many thought could not be achieved. However, today the whole of the human genome has been sequenced. The effort accelerated research into science. Since the sequencing of the genome the development of drugs, understanding the proteins involved in the interactions and the science of genetics, metabolomics and genome wide studies and omics studies have been able to flourish. Thus the case for mapping the protein interactions is a strong one. There can be no doubt that a Nobel Prize awaits the person who can achieve what would appear to be out of reach.
Citation: Khan R (2019) Developing High Sensitivity/Specificity Detection Systems for Studying Protein Interactions. J Proteomics Bioinform 12: 061-079. doi: 10.35248/0974-276X.19.12.498
Received: 11-Feb-2019 Accepted: 18-Apr-2019 Published: 26-Apr-2019 , DOI: 10.35248/0974-276X.19.12.498
Copyright: © 2019 Khan R. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.