Research Article - (2020)Volume 5, Issue 1
Background: At present, the diagnosis of gastric cancer lacks sufficient biomarkers. In this review, we will identify
and discuss the value of circulating free nucleic acids in the diagnosis of gastric cancer, in order to find a way to sort
out the diagnostic markers for gastric cancer.
Methods: A systematic search was performed on PubMed using the following keywords: (gastric cancer) and (blood or
plasma or serum) and (biomarker) and (DNA or RNA or cfDNA or cell-free DNA or RNA or CTC). and the
supplementary materials were added from Embase The Science Citation Index and The Cochrane. All the studies
contain biomarkers based on CTCs, DNAs, RNAs were reviewed and of which include sensitivity, specificity, and/or
AUC/ROC values were further discussed.
Results: 458 studies were searched and 87 contained biomarkers. Of these, 32 were prognostic, 12 were DNA, and
43 were RNA. Finally, 28 studies with complete data were included. The sensitivity and specificity ranged at
38.60%-95.50% and 44.30%-100%. The number of patients varied from 11 to 200, and the media is 60.
Additionally, 6 studies included independent validation cohort, and the media sensitivity and specificity were
83.95% and 80.35% respectively.
Conclusion: Although the sensitivity and specificity of biomarkers in current research are still insufficient, they also
show great potential value. Therefore, biomarkers with higher sensitivity and specificity require further exploration.
Gastric cancer; Circulating nucleic acids; Biomarker; Diagnosis
Gastric cancer has become the fifth common cancer disease and the third cause of cancer induced death in the world, with 1,033,701 new cases and 782,685 deaths in 2018 [1]. However, it is mainly distributed in East Asia, especially China [2]. Usually the early gastric cancer has no remarkable symptoms and it is often found to be a period of progress, therefore only a few patients were cured. There are still no relevant biomarkers for gastric cancer. According to the existing prognosis-related factors, most of them are age, gender, pathological type, and so on [3]. Currently, gastric cancer is diagnosed depending on gastro scopic biopsy as the gold standard.
Liquid biopsy is a major breakthrough in cancer research, especially with circulating blood. It provides a new perspective for noninvasive monitoring of tumors. Since Ashworth first reported cells found in the blood of tumor-dead patients similar to tumor cells, a large number of studies on circulating tumor cells in various tumors have emerged. With the development of next-generation sequencing technology, more and more researches are focused on nucleic acids based on circulating tumor cells. Nowadays, the detection of circulating tumor cells and circulating free nucleic acids have been proved having important clinical significance in the field of tumor diagnosis [4-6].
In this review, in order to provide some clues for finding diagnostic markers of gastric cancer, we mainly focus on the diagnostic value of circulating nucleic acids in gastric cancer, including RNA and DNA. To our knowledge, this is the first systematic review to discuss the diagnostic value of circulating nucleic acids in gastric cancer.
Four types of RNA have been found to be important for tumors, including miNRA, circRNA, LncRNA and mRNA. In addition, circulating cell-free DNA has also proven to be of value in the early diagnosis and prognosis of many tumors.
miRNA is a type of non-coding RNA, and its biological role is mainly in regulating gene expression [7,8] found that miRNAs in the circulatory system are relatively stable and have antiribonuclease properties. Therefore, it can be stored at room temperature for a long time and can be detected after repeated freeze-thaw cycles. There have been reported a large number of studies on the role of microRNAs in tumor growth, development, and metastasis [9].
circRNA is a special type of RNA, which was originally thought to be the result of mis-splicing during gene expression, but more and more studies have proved its role in tumor pathogenesis and progression [7,10]. Studies have found that low expression of circRNA in esophageal and colorectal cancer can inhibit tumor development [7,11,12]. In gastric cancer, some studies have found that the level of circRNA in tumor tissues has also increased, but there are still few studies.
LncRNA is a type of non-coding RNA with gene regulation effect over 200 nucleotides in length [13]. It can usually be detected in body fluids such as blood and urine of tumor patients. Studies have found that it has good diagnostic value in prostate cancer and esophageal cancer, and therefore has potential as a diagnostic marker [14,15].
As an intermediate product in gene expression, mRNA is detected and confirmed to play an important role in the blood of various tumor patients [16,17,18]. However, there is insufficient evidence as a diagnostic biomarker.
In 1948, Mandel first detected cell free DNA (cf-DNA) in human blood. In blood sample, cf-DNA is mainly derived from apoptotic nucleated cells. Apoptosis results in small, uniform DNA fragments, typically less than 180 bp in length, 3.6-5.0 ng / ml in normal human blood, and even more than 10 times in tumor patients [19]. As a branch of cf-DNA, circulating tumor DNA (ct-DNA) derived from tumor cells is one of the most extensive and in-depth research directions in the clinical management of tumor. Especially in evaluating the early detection and prognosis.
Search strategy
A systemic review of the literature was performed using PubMed on 20 March 2019 with the following keywords/MeSH words: (gastric cancer) and (blood or plasma or serum) and (biomarker) and (DNA or RNA or cf-DNA or cell-free DNA or RNA or CTC). The supplementary materials were added from Embase The science citation index and The Cochrane on 21 March.
Inclusion and exclusion criterion
The inclusion criteria includes: (1) all gastric cancer; (2) circulating nucleic acids (DNA and RNA) based on blood; (3) diagnostic biomarkers. While the exclusion criteria contains: (1) abstracts, reviews, comments, and letters; (2) duplicate studies; (3) proteins and tissues; (4) biomarkers for prognosis and treatment response; (5) incomplete data.
Data extraction and quality assessment
All the articles were filtered three times by three independent authors, and then the suitable studies were extracted by the corresponding author. According to the inclusion and exclusion criteria, the data about patients and control, type of biomarker, sensitivity, specificity, AUC were summarized by the first author. Afterwards the other author revised to confirm data was accurate.
Characteristic of studies
The inclusive flowchart is shown in Figure 1. A total of 449 studies were identified in the systematic database search. 94 studies were retrieved after assessment through titles and abstracts. Then 66 studies were excluded due to being irrelated with diagnosis or lacking of statistics. Finally, twenty-eight studies were included, among which, fourteen for miRNA, three for circRNA, six for LncRNA, one for mRNA, one for combination of miRNA and LncRNA, three for ct-DNA. A summary of the included studies is shown in Table 1 [20-47].
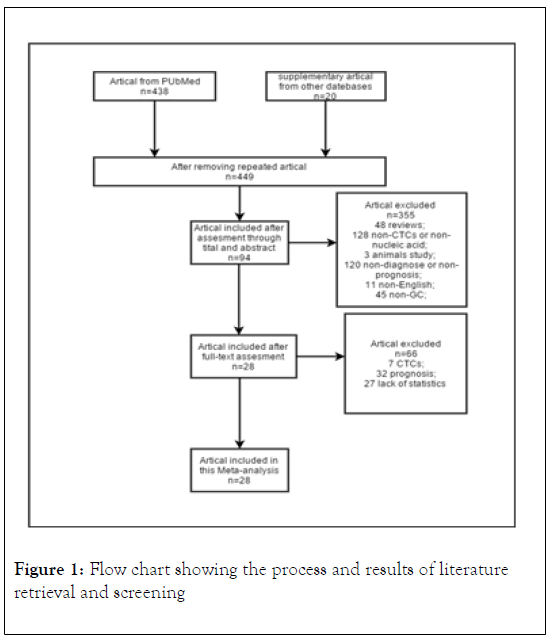
Figure 1: Flow chart showing the process and results of literature retrieval and screening.
| NO | Authors | Cancer type | Patients | Controls | Fluid | Method | Biomarkers | Sensitivity | Specificity | AUC | AUC 95% CI | Validation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | ||||||||||||
| 1 | Zhao et al. [45] | all GC | 11 GC | 17 normal | plasma | ddPCR | combination of miR-21, miR-93, miR-106a, miR-106b | 84.80% | 79.20% | 0.887 | 0.83–0.943 | YES |
| 2 | Li et al. [28] | all GC | 65 GC | 65 normal | plasma | qRT-PCR | miR 106b | 86.20% | 92.30% | 0.898 | 0.839-0.958 | NO |
| miR 25 | 87.60% | 76.90% | 0.817 | 0.738-0.897 | NO | |||||||
| miR 93 | 81.50% | 73.80% | 0.756 | 0.665-0.846 | NO | |||||||
| 3 | Qiu et al. [34] | all GC | 200 GC | 200 normal | plasma | qRT-PCR | miR-26a | 83.60% | 81.50% | 0.882 | 0.847-0.916 | YES |
| 4 | Liu et al. [31] | all GC | 80 GC | 70 normal | plasma | qRT-PCR | miR-940 | 81.25% | 98.57% | 0.966 | 0.940-0.992 | YES |
| 5 | Wu et al. [42] | all GC | 90 GC | 90 normal | Serum | qRT-PCR | miR-421 | 90% | 85.70% | 0.779 | 0.691-0.898 | NO |
| 6 | Wu et al. [43] | all GC | 50 GC | 50 normal | Serum | qRT-PCR | miR-21 | 88.40% | 79.60% | 0.912 | 0.869-0.968 | NO |
| 7 | Tsujiura M et al. [39] | all GC | 104 GC | 65 normal | plasma | qRT-PCR | miR-18a | 84.60% | 69.20% | 0.806 | NO | |
| 8 | Shin VY et al. [36] | all GC | 15 GC | 15 normal | plasma | qRT-PCR | combination of miR-627, miR-629, miR-652 | 86.70% | 85.50% | 0.941 | YES | |
| 9 | Wang et al. [41] | all GC | 50 GC | 47 normal | Serum | qRT-PCR | miR-223 | 81% | 78% | 0.85 | 0.780-0.930 | NO |
| miR-16 | 79% | 78% | 0.9 | 0.840-0.960 | NO | |||||||
| miR-100 | 71% | 58% | 0.71 | 0.610-0.820 | NO | |||||||
| 10 | Su et al. [37] | all GC | 82 GC | 65 normal | plasma | qRT-PCR | miR-18a | 80.50% | 84.60% | 0.907 | 0.860-0.953 | NO |
| 11 | Peng et al. [33] | all GC | 57 GC | 58 normal | Serum | qRT-PCR | miR-191 | 70.20% | 99.90% | 0.849 | 0.780-0.920 | NO |
| 12 | Li et al. [29] | all GC | 180 GC | 80 normal | plasma | qRT-PCR | miRNA-199a-3p | 80% | 74% | 0.837 | YES | |
| 20 precancerous lesions | ||||||||||||
| 13 | Valladares-Ayerbes M et al. [40] | all GC | 52 GC | 15 nromal | blood | qRT-PCR | miR-200c | 65.40% | 100% | 0.715 | 0.597-0.833 | NO |
| 14 | Li et al. [30] | all GC | 60 GC | 60 normal | plasma | qRT-PCR | miR-223 | 84.29% | 88.57% | 0.909 | 0.860-0.958 | YES |
| circRNA | ||||||||||||
| 15 | Huang et al. [25] | all GC | 60 GC | 60 normal | plasma | qRT-PCR | hsa_circ_0000745 | 85.50% | 45% | 0.683 | NO | |
| 16 | Chen et al. [20] | all GC | 104 GC | 104 normal | plasma | qRT-PCR | hsa_circ_0000190 | 41.40% | 87.50% | 0.6 | NO | |
| 17 | Sun et al. [38] | all GC | 45 GC | 17 normal | plasma | qRT-PCR | hsa_circ_0000520 | 82.35% | 84.44% | 0.897 | NO | |
| LncRNA | ||||||||||||
| 18 | Zong et al. [47] | all GC | 110 GC | 44 benign lesions | serum | RTFQ-PCR | lncRNA CTC497E21.4 | 81.82% | 75% | 0.848 | 0.794-0.901 | NO |
| 19 | Zhang et al. [44] | all GC | 162 GC | 110 normal | plasma | qRT-PCR | five lncRNA-based panel | 82% | 87% | 0.91 | 0.880-0.950 | NO |
| 28 precancerous lesions | 89% | 0.82 | 0.710-0.920 | NO | ||||||||
| 21 GIST | 86% | 0.8 | 0.680-0.910 | NO | ||||||||
| 20 | Jin et al. [26] | all GC | 100 GC | 110 normal | Serum | qRT-PCR | lncRNA HULC | 82% | 83.60% | 0.888 | 0.843-0.934 | NO |
| 21 | Hashad et al. [24] | all GC | 32 GC | 30 normal | plasma | qRT-PCR | LncRNA-H19 | 68.75% | 56.67% | 0.724 | NO | |
| 22 | Zhou et al. [46] | all GC | 70 GC | 70 normal | plasma | qRT-PCR | LncRNA-H19 | 82.90% | 72.90% | 0.838 | NO | |
| 23 | Gao et al. [22] | all GC | 20 GC | 20 normal | plasma | qRT-PCR | lncRNA-UCA1 | 89.20% | 80.30% | 0.928 | 0.888-0.968 | NO |
| miRNA and LncRNA | ||||||||||||
| 24 | Ghaedi et al. [23] | all GC | 62 GC | 40 normal | plasma | qRT-PCR | combination of H19, MEG3 and miR-675-5p | 89% | 85% | 0.927 | 0.850-0.960 | NO |
| mRNA | ||||||||||||
| 25 | Kang et al. [27] | all GC | 118 GC | 40 CAG / 58 normal | plasma | qRT-PCR | cell-free hTERT mRNA | 66% | 87% | 0.891 | NO | |
| DNA | ||||||||||||
| 26 | Chen et al. [21] | all GC | 104 GC | 20 normal | plasma | MSP | Zic1 promoter methylation | 60.60% | 61.40% | 0.61 | NO | |
| 50 GIN | 100% | 0.792 | NO | |||||||||
| 27 | Sakakura et al. [35] | all GC | 65 GC | 50 normal | Serum | RTQ-MSP | RUNX3 methylation | 95.50% | 62.50% | 0.865 | NO | |
| 28 | Park et al. [32] | all GC | 57 GC | 39 normal | plasma | qRT-PCR | MYC/GAPDH ratio | 38.60% | 100% | 0.816 | 0.732-0.899 | NO |
Table 1: Basic information for the 28 studies included.
We can find that in general, circulating nucleic acids show a sensitivity and specificity ranging from 38.60% to 95.50% and 44.30% to 100.00% in the diagnosis of gastric cancer. A total of 11-200 patients were included in the study, with a median of 60. In addition, six studies included independent validation cohorts (Table 2) with median sensitivity and specificity of 83.95% and 80.35%, respectively.
| Study details | Discovery cohort | Validation cohort | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Cancer type | Biomarkers | P | C | Sensitivity | Specificity | AUC | AUC | P | C | Sensitivity | Specificity | AUC | AUC |
| 95% CI | 95% CI | |||||||||||||
| Zhao et al. [45] | all GC | combination of miR-21, miR-93, miR-106a, miR-106b | 101 | 46 | Not given | 11 | 17 | 84.80% | 79.20% | 0.887 | 0.830–0.943 | |||
| Qiu et al. [34] | all GC | miR-26a | 80 | 80 | Not given | 200 | 200 | 83.60% | 81.50% | 0.882 | 0.847-0.916 | |||
| Liu et al. [31] | all GC | miR-940 | 30 | 30 | 60.00% | 96.67% | 0.896 | Not given | 80 | 70 | 81.25% | 98.57% | 0.966 | 0.940-0.992 |
| Shin et al. [36] | all GC | combination of miR-627, miR-629, miR-652 | 50 | 50 | Not given | 15 | 15 | 86.70% | 85.50% | 0.941 | Not given | |||
| Li et al. [29] | all GC | MiRNA-199a-3p | 30 | 30 | Not given | 180 | 100 | 80.00% | 74.00% | 0.837 | Not given | |||
| Li et al. [30] | all GC | miR-223 | 10 | 10 | 87.50% | Not given | 60 | 60 | 84.29% | 88.57% | 0.909 | 0.860-0.958 | ||
| miR-21 | 100.00% | Not given | 74.29% | 75.71% | 0.794 | 0.721-0.868 | ||||||||
| miR-218 | 87.50% | Not given | 94.29% | 44.29% | 0.743 | 0.663-0.824 | ||||||||
Table 2: Basic information on 6 studies in the validation cohort.
miRNA biomarkers
A total of 14 miRNA studies have demonstrated their value in the diagnosis of gastric cancer. Its sensitivity ranges from 65.40% to 94.29%, with a median of 82.55% specificity ranges from 44.29% to 100.00%, with a median of 79.40%. Twelve studies were performed on a single miRNA and two were performed on a combined miRNA. Six of these studies have a validation cohort.
In the study of Liu et al. [31] the level of miR-940 was reduced using a sample of 5 patients, and subsequently, a controlled study using 80 patients confirmed that the AUC reached 0.966, which is the highest in the current study. In the joint test, we found that its diagnostic value is more obvious than the single test. Zhao et al. [45] established the value of miR-21, miR-93, miR-106a, and miR-106b through a cohort of 147 people, and then verified the results using a cohort of 28 people. The sensitivity and specificity of the combination were improved compared with the four when independent, with an AUC of 0.887 when combined. Also, Shin et al. [36] Found 6 high levels of miRNAs in a control study of 123 gastric cancer patients and healthy people: miR-18a, miR-140-5p, miR-199a-3p, miR-627, miR-629 and miR-652. A 15-patient verification cohort was then used to find a combination of miR-627, miR-629, and miR-652, with sensitivity and specificity of 86.70% and 85.50%, respectively.
circRNA biomarkers
There are three circRNA-related studies, the most valuable of which is Sunet et al. [38] Their research showed that the sensitivity and specificity of hsa_circ_0000520 as a diagnostic marker for gastric cancer reached 82.35% and 84.44%, respectively. But the biggest problem is that their sample size is too small.
LncRNA biomarkers
A total of 6 studies have demonstrated the potential value of LncRNA in the diagnosis of gastric cancer. Five of them are for single LncRNA and one is for combined detection. The overall sensitivity of LncRNA is between 68.00%-89.20%, with a median of 81.91%; the overall specificity is between 56.67% -89.00%, and the median is 81.95%.
Studied from Hashad et al. [24] and Zhou et al. [46] revealed the value of LncRNA-H19. The former used 32 patients and controls with sensitivity and specificity lower than 70.00%, while the latter sample expanded to 70 cases In comparison, they all exceeded 70.00%. In the study of Zhang et al. [44] the combination of five lncRNAs (TINCR, CCAT2, AOC4P, BANCR and LINC00857) was found to have an AUC of 0.910 in the diagnosis of gastric cancer. In the second part, the combined test of the five also has some value in differential diagnosis. The AUCs that distinguish gastric cancer from precancerous lesions and gastric stromal tumors are 0.820 (95% CI: 0.710–0.920) and 0.800 (95% CI: 0.680–0.910).
mRNA biomarkers
There is only one study of mRNA. Kang’s et al. [27] found that the level of free hTERT mRNA was significantly increased in gastric cancer patients in a study of 118 patients with gastric cancer, 40 patients with chronic gastritis and 58 healthy controls (P<0.05). In the value of distinguishing gastric cancer patients, its sensitivity and specificity reach 66.00% and 87.00%, respectively, compared with CA19-9, CEA are significantly improved, so it may have important value in the diagnosis of gastric cancer.
Associated RNA biomarkers
Ghaedi et al. [23] found that LncRNA-H19, MEG3 levels were statistically different in 60 patients and 40 matched control groups (P<0.01), and then they detected the associated miRNA and found that miR-141-3p levels were also significantly reduced (P<0.05). Since then, the researchers have combined the three and found that they have a sensitivity of 88.87% and a specificity of 85.00% in distinguishing gastric cancer from controls, with an AUC of 0.927 (95% CI: 0.850-0.960). Therefore, the combination of these three may have some potential value for the diagnosis of gastric cancer. However, this study uses relevant RNA detection. If miRNA and LncRNA that are not very relevant can be used, then more research is still needed on whether the same results will be produced.
DNA biomarkers
There were 3 DNA studies, 2 of which were methylation of genes. In these studies, we found that the sensitivity and specificity have 'seesaw'-like results. That is one high and another low. Park et al. [32] used the ratio of MYC / GAPDH and found that its specificity can reach 100.00%, but unfortunately its sensitivity is only 38.60%.
There is currently no ideal biomarker in the diagnosis of gastric cancer. Although in this review we found 28 studies with potential, none of the studies were randomized controlled trials and their AUC values were the best. Therefore, the risk of bias increases. However, this is also an inevitable stage of early exploration.
In this review, various RNA and DNA molecules are reported. We found some common targets, including miR-21, miR-93, mi- R106, and LncRNA-H19. These targets have shown some value in both individual and combined detection. A new point is circRNA, which has been demonstrated by a total of 3 studies. Although the results are not very satisfactory, it also provides a new direction for gastric cancer diagnostic targets.
In this review we found that 20 studies extracted samples from plasma, while only 7 were extracted from serum, and 1 was extracted from whole blood. Although the concentration of nucleic acid in the serum is higher its advantages are greatly diminished due to the worse storage conditions of the serum. For whole blood, there are more interference factors and higher technical requirements, so its disadvantage is relatively obvious [48].
Because miRNA has a small molecular weight and is relatively stable, it is more accurate in detection. The number of studies included in the article is also the most, and the potential of miRNA is also confirmed from the side. miR-21, miR-93, and mi-R106 have been reported in different studies, but the results have significant differences. In the study of Zhao et al. [45] although the three were jointly tested, the results were still not ideal. From the validation queue, we can see that the joint detection results may be more stable. But unfortunately there is insufficient evidence for the detection of the combination, so whether the detection of the combination can provide better diagnostic value needs our further research.
From the study of miRNA combined with LncRNA, [23] we can see that the related miRNA and LncRNA have similar results in the diagnosis of gastric cancer, so the combined detection of these two has a certain feasibility to improve the value of detection. For both of them showing a certain value alone but not strongly related to each other, whether the combination still has sufficient value needs to be further explored.
Interestingly, we found that the circulating cell-free DNA has a "seesaw" effect as described above when used as a detection marker in gastric cancer. This may be due to the relatively poor stability of the DNA molecules, which can easily lead to loss during sample extraction. Therefore, its value as a diagnostic marker deserves further discussion.
As a diagnostic biomarker, another important factor is the differentiation from other diseases. Only one study has performed this step. In this study we found that the sensitivity and specificity of using a panel from five LncRNAs to identify gastric cancer and precancerous lesions were 68.00% and 89.00%, while for gastro intestinal stromal tumors were 68.00% and 86.00% [44].
Another note worthy result is that in the study by Zhang et al. [44] they found that the level of LncRNA diagnosed and detected before surgery was significantly reduced at 14 days after surgery (P=0.016). This may suggest that we can use this as a follow-up indicator of tumor progression for patients with high expression levels before surgery.
Although we saw many hopeful nucleic acid biomarkers in this review, these studies still have certain shortcomings, so we need more research to find more satisfactory nucleic acid biomarkers.
There is no acknowledge.
Citation: Zhang X, Zhou H, Wang T, Wu C, Zhao D (2020) Diagnostic Value of Circulating Nucleic Acids in Gastric Cancer: A Systematic Review. Immunogenet Open Access. 5:125.
Received: 14-Apr-2020 Accepted: 28-Apr-2020 Published: 05-May-2020 , DOI: 10.35248/igoa.20.5.125
Copyright: © 2020 Zhang X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.