Reproductive System & Sexual Disorders: Current Research
Open Access
ISSN: 2161-038X
ISSN: 2161-038X
Research Article - (2023)Volume 12, Issue 5
This study aimed to isolated and culture human Umbilial cord Mesenchymal Stem Cells (hUMSCs) in vitro by tissue block attachment to investigate their biological characteristics. The hUMSCs were cultured to passage 3, following which the induction experiments were performed in vitro and induced to melanocytes, neurons, and osteoblasts. The Reverse Transcription-Polymerase Chain Reaction (RT-PCR) results revealed that the expression of Nanog Homeobox (NANOG), Pou Class 5 Homeobox 1 (OCT4), 5'-nucleotidase ecto (CD73), CD44 molecule (CD44) in umbilical cord-derived mesenchymal stem cells were positive and population doubling time was 24.7 hrs. The differentiation properties of neurogenesis and osteogenic cells were confirmed by histological staining using toluidine blue and Alizarin red. Melanocytes were detected by RT-PCR, quantitative PCR (qPCR), and immunofluorescence staining. After 28 days of differentiation, the cells exhibited a typical morphology such as bipolar or tripolar cells with slender dendrites. The immunofluorescence staining showed that the differentiated cells expressed Microphthalmia Transcription Factor (MITF) and Tyrosinase (TYR), the expression of MITF, TYR and KIT proto-oncogene receptor tyrosine Kinase (KIT) from 0 d to 28 days by qPCR was significantly different and the expression of marker genes MITF, SOX10, KIT by RT-PCR were positive. The results demonstrated that the cells isolated from human umbilical cord were mesenchymal stem cells, and hUMSCs had multidifferentiation potentialities. hUMSCs could also differentiate into melanocytes in vitro, providing reliable sources of melanocytes for treating vitiligo future.
Cell culture; Melanocyte cell; human Umbilical Cord-derived Mesenchymal Stem Cells (hUMSCs)
Mesenchymal Stem Cells (MSCs) are pluripotent stem cells existing in multiple tissues, they have strong self-renewal and differentiation abilities. MSCs can be induced to differentiate into cells of various embryonic layers, such as osteoblasts, nerve cells, cardiomyocyte, keratinocytes, and adipocytes [1-4]. Therefore, these cells have significant importance to gene therapy, regenerative medicine, tissue engineering and are considered a particularly attractive cell type for cell-based therapies in human [5-8].
Compared with traditional donor cells and fetal fibroblast cells, MSCs with rapid proliferation and anti-aging abilities can also shorten nuclear reprogramming time. Compared with embryonic stem cells, adult stem cells cannot be easily cultured in large numbers, but their differentiation is relatively stable. At present, many types of adult stem cells have entered the clinical research stage [9-11].
Human umbilical cord is a new source of MSCs that exhibit a high degree of self-renewal capacity and multi-differentiation potential. human Umbilical Cord Derived Mesenchymal Stem cells (hUMSCs) have a wider range of collection sources than BMSCs or ESCs, and can be easily collected with fewer ethical constraints. As an alternative source of MSCs, hUMSCs have promising clinical application prospects [12,13].
In this study, human Umbilical Cord Derived Mesenchymal Stem Cells (hUMSCs) were isolated and cultured, and their biological characteristics were studied. Then, hUMSCs were induced to differentiate into osteoblasts, neuronal and melanocyte cells and were identified based on the organizational and the gene level. The differentiation properties of neurogenesis and osteogenic cells were confirmed by histological staining using toluidine blue and Alizarin red.
In particularly, we induced umbilical cord mesenchymal stem cells into melanocytes. Vitiligo, a disease that is characterized by loss of melanocytes in the skin. Vitiligo affects 0.1% to greater than 8% of population worldwide. Autologous melanocyte transplantation is effective for treating stable vitiligo and is a promising cell therapy. The procedure is not widely used because human adult melanocytes are difficult to culture and amplify large scale in vitro. In the present, which can induce melanocytes from adipose MSC-derived Muse cells [14]. These studies suggest that induction of melanocytes from hUMSCs can clinical application and in vitro study of melanocyte differentiation.
All chemical reagents and culture media were cell culture grade and obtained from Sigma Chemicals (America), while other sources were labelled. The plastic ware was obtained from Corning (America).
Umbilical cord collection
The umbilical cord tissue of healthy male infants without hepatitis, Acquired Immunodeficiency Syndrome (AIDS), and other infectious diseases was taken from the operation room of the Hohhot First Hospital. The ethics committee approved the study, and the family members provided informed consent. The tissue was collected and stored in the phosphate buffer containing 1% penicillin and streptomycin sulfate. The samples were immediately transported to the laboratory for father processing.
Cell isolation and culture
The umbilical cord in the clean bench was taken out, put into the Dulbecco’s Phosphate Buffered Saline (D-PBS, Gibco) with 1% penicillin/streptomycin (100 U/mL and 100 μg/mL; HyClone), and cleaned three times to remove the blood as much as possible. Alcohol was used for umbilical cord disinfection, following which the cord was placed in D-PBS with 1% penicillin/streptomycin for cleaning three times to remove the residual alcohol. The umbilical vein cavity was cut to remove two arteries and one vein using ophthalmic scissors. The umbilical cord tissue was divided into several 2-cm-long parts, and the Wharton’s Jelly (WJ) was separated. The WJ was cut into 0.5 to 1 cm2 segments and several crisscross scratches were made on its surface, but it was not cut through the tissue block. The tissue block was placed in the culture dish. The tissue block was cultured with Dulbecco’s Modified Eagle’s Medium (DMEM)-F12 (Gibco, Rockville) supplemented with 10% Fetal Bovine Serum (FBS, Gibco) and 0.1% penicillin/streptomycin at 37°C in a 5% CO2 humidified incubator. The cells crawled out after 7-9 days. When the adherent cells reached 80%-90%, they were digested with trypsin (0.05% trypsin-EDTA, Gibco-Invitrogen) and then frozen or subculture. The culture medium was changed every 2-3 days.
Cryopreservation and reconstitution
The cell cryopreservation was performed as described by Guan and Hao [13,15]. When the cells were frozen, the cell density of each tube was 1 × 106 cell/mL. The cryopreserved solution consisted of a mixture of the following reagents: 70% DMEM/F12, 20% Fetal Bovine Serum (FBS), and 10% dimethyl sulfoxide (Shanghai Chemical, Shanghai, China). When the cells were resuscitated, the cells were recovered by shaking the cryovial in 37°C water baths for 1 min. The contents in the tube were transferred into a 1.5 mL centrifuge tube (Nunc) and centrifuged at 1400 rpm for 5 min. The supernatant was removed, mixed with culture solution, and then added to a new culture dish in a 1:3 ratio, followed by culturing at 37°C with saturated humidity and 5% CO2.
Cell doubling method
After resuspension, the hUMSCs were cultured into a 24-well plate to achieve a cell density of 1 × 104 cells/well. The cells in three wells were counted every day, and the average value was taken. The growth curve was drawn after 8 days of calculation. The population-doubling time was calculated as t [lg2/ (lgNt-lgN0)] (N0 and Nt represent the cell counts after inoculation and t the hours after the start of culturing, respectively).
Reverse Transcription Polymerase Chain Reaction (RTPCR)
The stem cell-specific gene expression in hUMSCs isolated and cultured in vitro was analyzed by RT-PCR [16]. Briefly, total RNA was isolated from hUMSCs at passage 3 using RNAiso Plus (TaKaRa, Otsu, Japan). Then, a 10 µL reverse transcription system was prepared following the instructions on the reverse transcription kit: Enzyme (PrimeScript RT Master Mix, TaKaRa, Japan), 2 µL; and RNA template, 500-1000 ng. The system was supplemented with enzyme-free water to 10 µL. The conditions for RT were as follows: 37°C for 15 min, 85°C for 5 s, and 4°C till the reaction was completed. The synthesized cDNA was stored at -20°C.
A 20 µL system was prepared according to the instructions of PCR kit: Enzyme (Premix Taq, TaKaRa, Japan), 10 µL; cDNA template, 50-100 ng; and primer, 2 µL. The conditions for amplification were as follows: 35 cycles each consisting of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, elongation at 72°C for 30 s, final extension at 72°C for 5 min, and at 4°C till the reaction was completed. The PCR primers and the reaction conditions are summarized in Table 1. After the cDNA was amplified, agarose gel electrophoresis was performed and visualized under a gel documentation system (Gene genius, England).
| Gene | Primer sequence | Product size (bp) | Annealing temperature (?) | Source | Accession no. |
|---|---|---|---|---|---|
| OCT4 | CGAGAACCGAGTGAGAGGCAAC A(F) | 298 | 59.5 | Homo sapiens | NM_001173531.3 |
| CGAGGAGTACAGTGCAGTGAAGTG(R) | |||||
| NANOG | TACCTCAGCCTCCAGCAGATGC(F) | 201 | 61.5 | Homo sapiens | NM_001297698.2 |
| CCAGGTCTGGTTGCTCCACATTG(R) | |||||
| GAPDH | GGCACCGTCAAGGCTGAGAAC (F) | 381 | 58 | Homo sapiens | NM_001256799.3 |
| GGTGGCAGTGATGGCATGGAC(R) | |||||
| CD44 | ACGFAGCAGAGTAATTCTCAGAGC(F) | 319 | 59.5 | Homo sapiens | NM_000610.4 |
| GGTGGAATGTGTCTTGGTCTCCTG(R) | |||||
| CD73 | GCTCGGCTCTTCACCAAGGTTC(F) | 215 | 61 | Homo sapiens | NM_001204813.2 |
| TTGAGGAGTGGCTCGATCAGTCC(R) | |||||
| MITF | GGCACCARCACCTTCAACAACAAC(F) | 214 | 61 | Homo sapiens | NM_000248.4 |
| GCTCCGTCTCTTCCATGCTCATAC(R) | |||||
| KIT | GCACCGAAGGAGGCACTTACAC(F) | 219 | 61 | Homo sapiens | NM_000222.3 |
| CTGGCAGTACAGAAGCAGAGCATC(R) | |||||
| TYR | GAAGGCACCGTCCTCTTCAAGAAG(F) | 223 | 60.5 | Homo sapiens | NM_000372.5 |
| GACCAGARCCGACTCGCTTGTTC(R) | |||||
| SOX10 | GCAAGGTCAAGAAGGAGCAGCAG(F) | 109 | 62 | Homo sapiens | NM_006941.4 |
| ACCAGCGTCCAGTCGTAGCCC(R) |
Note: Base pair (bp); source for all primers is Homo sapiens arise from NCBI.
Table 1: Details of primers used for gene expression through RT-PCR and qPCR.
Quantitative real-time PCR
The expression of targeted genes in melanocyte cells was measured using a qPCR assay on Roche LightCycler 480 II Time PCR System (Basel, Switzerland). The PCR primers and the reaction conditions are summarized in Table 1. Total RNA was extracted with Trizol reagent and quality controlled with Agilent 2100 Bioanalyzer (America). cDNA reverse transcription was performed by using Synthesis SuperMix (TaKaRa, Otsu, Japan). The PCR system was 20 µL, including 10 µL of SYBR Green qPCR Master Mix (TaKaRa, Ostu, Japan), PCR forward primer, PCR reverse primer, 1 µL of template cDNA, and ddH2O, with the following reaction conditions: initial denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 3 s and annealing and extension at 60°C for 30 s. GAPDH was used as reference gene, and the 2-ΔΔCT method was used to analyses the relative mRNA expression. Each experiment was conducted in triplicate.
Induced differentiation in vitro
The cells were cultured in six-well plates for passage 3. When the cell reached 70%-80%, they were added to the osteogenic induction fluid for osteogenic induction. A 50-mL system of an osteogenic induction medium was prepared as follows: 10% FBS, 10 mmol/L β-glycerophosphate, 20 nmol/L dexamethasone, and 50 μg/mL sodium 2-phosphate ascorbate in DMEM-F12). The cell differentiation was assessed morphologically, and they were stained with alizarin red and von Kossa [2].
To induce neural cells, hUMSCs were exposed to neuronal preinduction media (a 50-mL system was prepared as follows: 20% FBS and 10-3 mol/L β-mercaptoethanol (Shanghai Chemical, Cat. #386) in DMEM). After 24 h, the cells were washed twice with D-PBS and placed in 5×10-3 mol/L β-mercaptoethanol in DMEM without FBS for 6 h [17]. The cells were stained with toluidine blue and observed under the microscope.
To induce melanocyte cells differentiation, a 50 mL system was prepared as follow: 10 mL MCDB201 medium; 15 mL DMEM; 2.5 mm dexamethasone 1 µL; 0.3 mg/ml cholera toxin 0.28 µL; 1 mm TPA 25 µL; insulin transferrin selenium 500 µL; linoleic acid aluminum from bovine serum aluminum 500 µL; L-ascorbic acid 0.0009 g; Glutamax 500 µL; 10 μg/ml SCF 250 µL; 100 µm ET-3 50 µL; 4 μg/ml bFGF 50 µL; 25 ml L-wnt-3a cell suspension and store in a dark place at 4° after sub-packaging. The solution was changed every 2 days, and the induction of cells ended after 28 days. The cell differentiation was assessed morphologically (Table 1).
Staining methods
• Alizarin red staining. Medium was aspirated from culture wells. The cells were fixed in 4% paraformaldehyde for 1 h, washed twice with water, and incubated with Alizarin red solution at room temperature for 30 min. After incubation, stain was removed. The cells were washed four times thoroughly with water, finally, ~1 mL of water was left to prevent cells from drying out. The cells were visualized under the phase contrast microscope.
• Toluidine blue staining. The cells were rinsed two times with PBS, then fixed in 4% formalin and stained in a toluidine blue working solution for 2-3 min. Next, they were washed in running tap water for 2 min, rinsed in distilled water, and observed under the microscope.
Immunofluorescence
When the cell reached of 70%-80%, the culture solution was fixed with 4% paraformaldehyde. After 10 min, it was sealed with 1% bovine serum albumin at room temperature for 30 min. Futher, it was incubated with MITF (Mouse anti-human monoclonal antibodies to MITF), TYR (Mouse anti-human monoclonal antibodies) antibody (1:1000, Abcam, Cambridge, UK) at 4°C overnight and then with FITC (Goat Anti-Mouse IgG H&L)-labeled secondary antibody (1;500, Abcam, Cambridge, UK) at 25°C for 50 min. 4’,6-Diamidino- 2-Phenylindole (DAPI) was used to label the nucleus.
Isolation and culture of hUMSCs
The tissue block was placed in a 60 mm2 culture dish. The fluid was not changed for the first 3 days and then changed every 2 days. After 7-9 days, fibroblast-like cells crawled out of the bottom of the dish (Figure 1A). Most of the cells were typical spindle-like cell; about 12 days later, they reached 80% confluence. After three generations of culture, the cell morphology remained consistent, showing a long spindle shape (Figure 1B). When the cells converged to 80%-90%, they were frozen and stored in liquid nitrogen for long-term storage and later use.
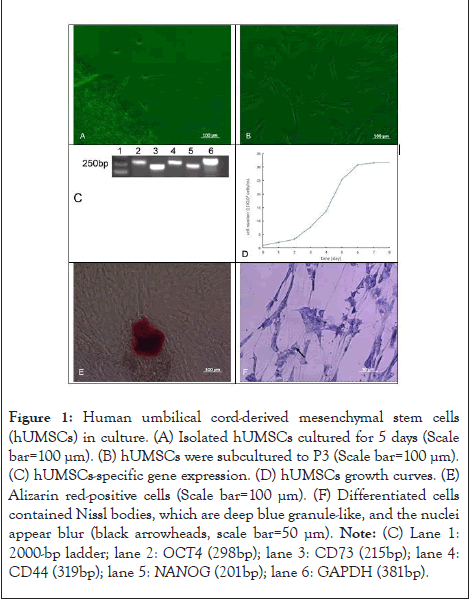
Figure 1: Human umbilical cord-derived mesenchymal stem cells
(hUMSCs) in culture. (A) Isolated hUMSCs cultured for 5 days (Scale
bar=100 µm). (B) hUMSCs were subcultured to P3 (Scale bar=100 µm).
(C) hUMSCs-specific gene expression. (D) hUMSCs growth curves. (E)
Alizarin red-positive cells (Scale bar=100 µm). (F) Differentiated cells
contained Nissl bodies, which are deep blue granule-like, and the nuclei
appear blur (black arrowheads, scale bar=50 µm). Note: (C) Lane 1:
2000-bp ladder; lane 2: OCT4 (298bp); lane 3: CD73 (215bp); lane 4:
CD44 (319bp); lane 5: NANOG (201bp); lane 6: GAPDH (381bp).
Characterization of hUMSCs
The RT-PCR results showed that the specific genes OCT4, CD73, CD44 and NANOG of hUMSCs were expressed positively (Figure 1C). The cell growth curves were drawn for hUMSCs at P3. The results showed that the growth curve had a typical S-shape. The growth was slow from the first to the second day, increased from the third to the sixth day, and reached the platen stage from the seventh day. The average PDT of the hUMSCs was calculated to be 24.7 hrs (Figure 1D). It showed that the cells quality is high, genetic, and stable, can be used in subsequent experiments (Figure 1).
Osteogenic differentiation of hUMSCs
When osteoblast induction solution was added, the cells began to show morphological changes on the 12th day, and alizarin red staining was performed 21 days later (Figure 1E). Alizarin red staining showed positive expression, indicating that hUMSCs could be induced to differentiate into osteoblasts in vitro.
Neural differentiation of hUMSCs
Following nerve induction and differentiation, Nissel bodies were found in differentiated cells with blue granules and dark blue nuclei after toluidine blue staining (Figure 1F). The results showed that hUMSCs could be induced to differentiate into neural cells in vitro.
Melanocytes cell differentiation of hUMSCs
The cells differentiated into melanocytes in 0 to 28 d (Figure 2A- 2C). The cells morphology has changed at 14 d (Figure 2B). The cells changed from fibroblast-like to bipolar or tripolar cells with slender dendrites, and the appearance was similar to that of normal melanocytes (Figure 2C). Immunofluorescence staining was performed to confirm the phenotype of these differentiated cells, revealing that they expressed specific markers MITF and TYR [18-20]. The RT-PCR of the cultured differentiated cells indicated positive expression of MITF, KIT and SOX10 (Figure 2D). The relative expression of MITF, KIT, and TYR increased in differentiated cells with the increase in the culture time compared with that in the control group (Figure 2E). MITF and TYR were expressed in the differentiated cells, as detected by Immunofluorescent staining (Figure 3A-3F and 4A-4F). This proved that hUMSCs successfully differentiated into melanocytes and expressed marker genes (Figure 2-4).
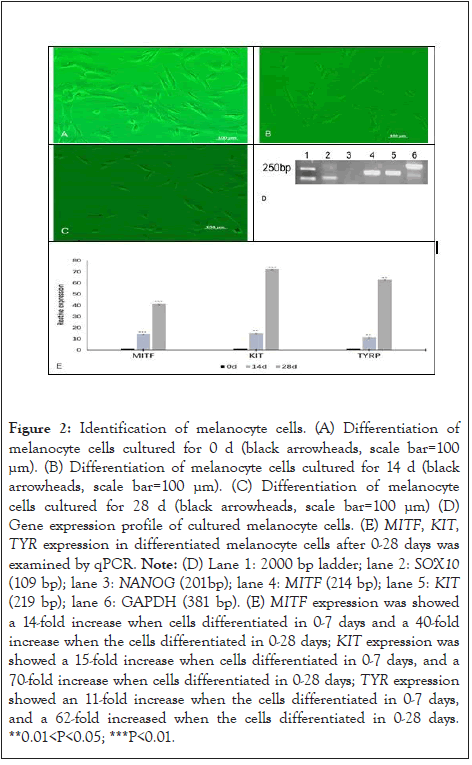
Figure 2: Identification of melanocyte cells. (A) Differentiation of melanocyte cells cultured for 0 d (black arrowheads, scale bar=100 µm). (B) Differentiation of melanocyte cells cultured for 14 d (black arrowheads, scale bar=100 µm). (C) Differentiation of melanocyte cells cultured for 28 d (black arrowheads, scale bar=100 µm) (D) Gene expression profile of cultured melanocyte cells. (E) MITF, KIT, TYR expression in differentiated melanocyte cells after 0-28 days was examined by qPCR. Note: (D) Lane 1: 2000 bp ladder; lane 2: SOX10 (109 bp); lane 3: NANOG (201bp); lane 4: MITF (214 bp); lane 5: KIT (219 bp); lane 6: GAPDH (381 bp). (E) MITF expression was showed a 14-fold increase when cells differentiated in 0-7 days and a 40-fold increase when the cells differentiated in 0-28 days; KIT expression was showed a 15-fold increase when cells differentiated in 0-7 days, and a 70-fold increase when cells differentiated in 0-28 days; TYR expression showed an 11-fold increase when the cells differentiated in 0-7 days, and a 62-fold increased when the cells differentiated in 0-28 days. **0.01<P<0.05; ***P<0.01.
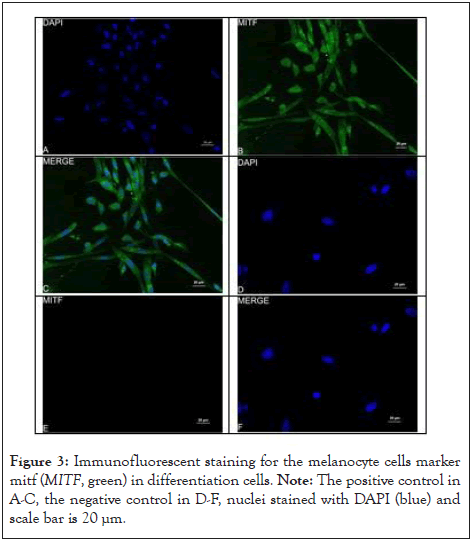
Figure 3: Immunofluorescent staining for the melanocyte cells marker mitf (MITF, green) in differentiation cells. Note: The positive control in A-C, the negative control in D-F, nuclei stained with DAPI (blue) and scale bar is 20 µm.
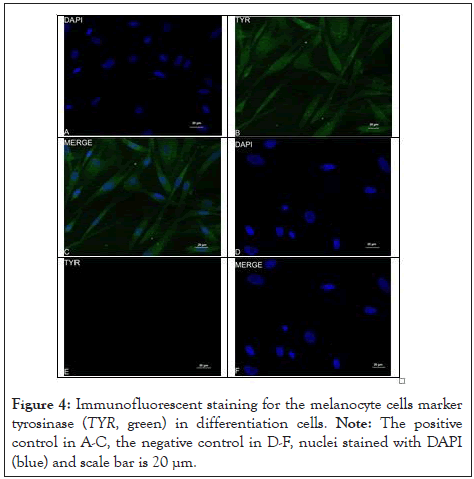
Figure 4: Immunofluorescent staining for the melanocyte cells marker tyrosinase (TYR, green) in differentiation cells. Note: The positive control in A-C, the negative control in D-F, nuclei stained with DAPI (blue) and scale bar is 20 µm.
In this study, hUMSCs were isolated and cultured in vitro and induced to differentiate. The induction solutions of osteoblasts, neurons, and melanocytes were added. The cells induced to differentiate were successfully obtained through principal component analyses. It proved that hUMSCs could be induced to differentiate into many kinds of cells in vitro [8]. The self-renewal and multi-differentiation abilities of hUMSCs laid the foundation for cell therapy.
In this study, hUMSCs were obtained by adherent culture of healthy male infant’s umbilical cord. hUMSCs showed typical fibrous and long spindles, the results of cell viability prior to freezing and after cell recovery indicated that freezing had little influence on the viability of the hUUMSCs. The growth curves of hUMSCs with an S-shape. Since the exponential phase, the average PDT for P3 MSCs was 24.7 h. A few transcription factors, like Oct4, Nanog, Sox2, their expressions were used to characterize stem cells in various species. In this study, the expressions of Oct4, Sox2 and Nanog were studied to characterize the stemness of P3 hUMSCs. RT-PCR shows that these genes were positively expressed, which agrees with, who reported the expression of these genes in porcine BM-derived cells [21].
hUMSCs have self-renewal and high differentiation capacity. In this study, different types of defined media were added to the cultured cells, and the hUMSCs were induced to differentiate separately into osteoblasts. Alizarin red staining was positive, as reported earlier [22- 24]. These results suggest that the hUMSCs can differentiate into osteoblasts.
β-mercaptoethanol could support the viability and differentiation of fetal mouse brain neurons and was used as an effective inducer of neural differentiation in MSC [25-27]. β-mercaptoethanol also induced dramatic modifications of cellular shape and the expression of neural marker NeuN with 5 h [28,29]. This study found that differentiated cells contain Nissl bodies which are deep blue granule-like, and the nuclei appear blur, indicate hUMSCs also could differentiate into neural cells in serum-free conditions.
The induction and differentiation of melanocytes requires many cytokines, and the factors in vitro affect their growth [30]. The Microphthalmia-Associated Transcription Factor (MITF) is the master regulator of melanocyte differentiation, development, and survival [31]. It plays a central role in the complex network of interacting genes regulating the migration, survival, and proliferation of melanocytes [32-34]. In this study, the expressions of MITF, TYR and KIT were studied to characterize the melanocytes. RT-PCR shows that these genes are positively expressed, immunofluorescent staining for the melanocyte cells marker genes MITF, TYR in differentiation cells were positive, and MITF, KIT, TYR expression in differentiated melanocyte cells examined by qPCR were significant difference. Indicate hUMSCs also could differentiate into melanocyte cell in vitro.
We found that hUMSCs can be cultured in vitro, exhibit good growth, and have high proliferation ability. The expression of pluripotency marker genes showed that these cells have the characteristics of stem cells and could be induced to differentiate into melanocyte cells. Next, we will make melanin in vivo using our human induced pluripotent stem cell-derived melanocytes.
Z.C.X wrote the main manuscript text and prepared Figures 3-4, G. preppared Figures 1-2, Z.Y.R, C.J.W, L.C.X and Z.H.M provide the idea about article. W.S.C and L.L provide the Materials. All authors reviewed the manuscript.
This study was supported by a Grant-in-Aid for the major project of the Inner Mongolia Natural Science Foundation (2020ZD0003), and Inner Mongolia Key Laboratory of Biomanufacture.
The authors declare no conflict of interest.
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee. Written informed consent was obtained from individual or guardian participants.
All data generated or analysed during this study are included in this published article.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Zhang Y, Zhu C, Guo Y, Liu L, Cao J, Liu C, et al. (2023) Differentiation of Human Umbilical Cord-derived Mesenchymal Stem Cells to Melanocyte, Osteogenic and Neural Cells In Vitro. Reprod Syst Sex Disord.12:382.
Received: 21-Sep-2023, Manuscript No. RSSD-23-27110; Editor assigned: 25-Sep-2023, Pre QC No. RSSD-23-27110 (PQ); Reviewed: 09-Oct-2023, QC No. RSSD-23-27110; Revised: 16-Oct-2023, Manuscript No. RSSD-23-27110 (R); Published: 23-Oct-2023 , DOI: 10.35248/2161-038X.23.12.382
Copyright: ©2023 Zhang Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.