Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2011) Volume 4, Issue 12
Calculating Biological functionalities of proteins and presenting them numerically is an approach that will benefit the designing of drugs and vaccines. For example, potency of vaccines is known to be measured in terms of specificity, which is determined by bio-recognition (affinity), and sensitivity. Calculating the bio-recognition of peptides employed in the design of vaccines by means of procedures like the Digital Signal Processing (DSP) techniques rather than clinical experimentations not only gives room for the manipulation of the amino acids sequences of the peptides but also helps determine the degree of specificity of the antibody generated, hence the potency of the vaccine produced. This also provides opportunity for optimatization of the peptides for the desired biological characteristics. Also comparing the potency of peptide-based drugs through calculation of their biological functions is a faster, easier and resource-saving approach to pharmacotherapeutic investigations. In this study, two DSP techniques are fully explained and demonstrated. They are Resonant Recognition Model and Informational Spectrum Method. They are employed in the calculation of some physiological characteristics of Plasmodial peptides (P18 and P32), which are still being study for possible use as materials for the designing of anti-Malaria vaccines. Furthermore, the approaches are utilised to assess the pharmacological activities of two Fusion inhibitors (Enfuvirtide and Sifuvirtide). Enfuvirtide is currently in use for the management of anti-HIV/AIDS while Sifuvirtide is still being studied. Our calculated results demonstrate strong correlation with the preliminary clinical findings. They also seem to suggest that presenting biological characteristics in numerical terms is an easier and more rational approach to designing drugs and vaccines as it save resources and time unlike clinical experimentations. The methods also appear to help simplify the manoeuvring of the protein residues, which are employed in the designing and development of drugs and vaccines in order to obtain maximal biological characteristics.
Keywords: Digital signal processing; Fusion peptide; Informational spectrum method; Resonant recognition model
Digital Signal Processing techniques are analytic procedures, which decompose and process signals in order to reveal information embedded in them [1]. The signals may be continuous (unending) or discrete such as the protein residues. Digital Signal Processing techniques have helped analyse protein interactions [2] and made biological functionalities calculable including drug resistance [3]. This is in contrast to Bioinformatics methods like Multiple Structural Alignment (MSA), which dwells on homology and as such predict protein functionalities.
In these approaches, protein residues are first converted into numerical sequences (signals) using one out of over 600 available amino acids parameters that are responsible for the biological functionality. These numerical sequences (signals) are then processed by means of Discrete Fourier Transform (DFT) to present the biological characteristics of the proteins in the form of Informational Spectrum. This procedure is called Informational Spectrum Method (ISM) [4-6].
A version of the ISM, which engages amino acids parameter called Electron-Ion Interaction Potential (EIIP) is referred Resonant Recognition Model (RRM). In this procedure, biological functionalities are presented as Spectral Characteristics [2]. This physico-mathematical process, which is based on the fact that bio-molecules with same biological characteristics recognise and bio-attach to themselves when their valence electrons oscillate and then reverberate in an electromagnetic field [2].
In this study, detailed explanation of Resonant Recognition Model and Informational Spectrum Method (ISM) are provided. Furthermore, demonstration of these steps is made using one out of over 600 amino acids parameters, and two Plasmodial peptides (P18 and P32) [4]. Thereafter, the techniques are used to calculate biological functionalities of peptide-based anti-HIV /AIDS drugs (Enfuvirtide and Sifuvirtide) and the two Plasmodial peptides (P18 and P32) which are presented numerically in order to study their functionalities.
Resonant Recognition Model (RRM): Background
Resonant Recognition Model (RRM) is a Digital Signal Processingbased technique, which recognizes protein primary structures or physiological functionalities as protein residues represented by numbers that are assigned from the Electron-ion Interaction Pseudopotenatial (EIIP) parameter [2]. RRM involves four steps, which are fully discussed below.
The steps include:
Step 1: Conversion of the Protein Residues into Numerical Values Electron-ion Interaction Pseudopotenatial (EIIP) Parameter: There are 20 essential amino acids constituents (protein residues) [7]. Biological interactions have been studied in relation to the behaviour of these 20 amino acids constituent of proteins. As a result, the level of participation by these protein residues in over 565 protein-protein interactions that characterise biological functionalities have been derived as Amino Acids Parameters (AAPs) and deposited in databases like www.genome.jp/aaindex/ [8] and literatures such as [9]. AAPs describe physiochemical characteristics such as Hydropathy [10]; Hydrophobicity [11,12]; Hydrophilicity [13,14]; and Structural features including Alpha-Helix [15,16], and Beta [16] conformations.
Electron-Ion Interaction Potential (EIIP) (Table 1) is one of the amino acid parameter. The Resonant Recognition Model (RRM) engages EIIP amino acids parameter. Proteins molecules are known to be sequences of amino acids with unrestricted electrons and charges [17]. These charges elicit short-lived polarization of the side-chain groups and result in electromagnetic oscillation between some parts of the protein molecule [18]. These swinging in the protein molecules interfere with one another [19,20]. During oscillation, molecules, which share same biological characteristics, are found to resonate at the same frequency leading to amplified attraction (affinity) [18-20]. This reverberation, which arises from electromagnetic oscillation between bio-molecules (electromagnetic resonance) [2], is called the Electron- Ion Interaction Potential (EIIP) [19,21]. RRM which engages EIIP and therefore determines biological characteristics of the protein residues in terms of bio-recognition (specificity) and binding interaction (affinity) [2]. Bio-recognition and bio-attachment are the first two steps in molecular interactions. By means of DSP, the biological functionalities of the peptide-based anti-HIV/AIDS drugs and Plasmodial peptides are calculated. This is in an attempt to find out if this rational approach is realizable and beneficial. Plasmodial peptides are prospective starter materials for Malaria vaccine design peptides.
| Amino Acid | Three Letter Code | Letter Code | EIIP |
|---|---|---|---|
| Alanine | Ala | A | 0.0373 |
| Arginine | Arg | R | 0.0959 |
| Asparagine | Asn | N | 0.1263 |
| Aspartic acid | Asp | D | 0.0036 |
| Cysteine | Cys | C | 0.0829 |
| Glutamine | Gln | Q | 0.0761 |
| Glutamic acid | Glu | E | 0.0058 |
| Glycine | Gly | G | 0.0050 |
| Histidine | His | H | 0.0242 |
| Isoleucine | Iso | I | 0.0000 |
| Leucine | Leu | L | 0.0000 |
| Lysine | Lys | K | 0.0371 |
| Methionine | Met | M | 0.0823 |
| Phenylalanine | Phe | F | 0.0946 |
| Proline | Pro | P | 0.0198 |
| Serine | Ser | S | 0.0829 |
| Threonine | Thr | T | 0.0941 |
| Tryptophan | Trp | W | 0.0548 |
| Tyrosine | Tyr | Y | 0.0516 |
| Valine | Val | V | 0.0057 |
Table 1: Amino Acids Parameter: Electron-ion Interaction Pseudopotenatial (EIIP)
Step 2: Zero-padding/Upsampling: In some cases, proteins that are to be analyzed by means of RRM may have unequal residues. Signal Processing techniques demand that the window length of all proteins be the same [1,28]. Such unequal window lengths can be found in Plasmodial peptides P18 and P32 being used in this study as shown in Table 2. P18 has 16 amino acids compositions while P32 has the largest protein length (N), which is 32. Therefore, prior to the decomposition of the numerical signals by means Fourier Transform (FT), the sequences are to be brought to same sequence (window) length with largest protein residue (N). In the case of peptides P18 and P32, 14 zeros are added to peptide P18. This is called zero-padding or up-sampling [22]. Zero-padding operation is usually used to improve the visual clarity of the spectrum. It does not improve the quality of the spectral results. However, zero-padding has been claimed to constitute error [23].
| Peptide Identity | Peptide |
|---|---|
| P18 | EWSPCSVTCGNGIQVRIK |
| P32 | IEQYLKKIKNSISTEWSPCSVTCGNGIQVRIK |
Table 2: Peptides for the Demonstration of RRM and ISM.
Step 3: Processing of the Numerical Sequences (Signals) using Fourier Transform(FFT): Fourier Transform is a mathematical operation that converts one signal to another without altering the information contained in the signal [1]. This is in an attempt to disclose hidden information. Because proteins are amino acids in compartments (digitalized), Discrete Fourier Transform (DFT) is employed in their analysis [1]. Though DFT is known to be the most straight-forward mathematical procedure [1], it is found to be inefficient.
As a result, a more efficient, rational and faster algorithm, which executes DFT faster without altering the results, is being employed [24]. It is called Fast Fourier Transform (FFT), and is represented as:
y=abs(fft(x)) (1)
RRM-based decomposed signals results in three sets of values namely imaginary, real and absolute. The plots of these values describe the Spectral Characteristics (SC) of proteins. The Absolute values have been exploited in assessing biological functionalities of proteins. However, Imaginary and Real values have recently been found to reveal some biological characteristics [25]. The y-axis (Amplitude) symbolizes the bio-recognition (specificity) and binding (affinity) of each protein residue while the x-axis (Frequency) determines the position of the interaction.
Step 4: Cross-spectral Analysis: In order to identify the common biological relationship amongst proteins, point-wise multiplication of the Spectral Characteristics of the protein residues are applied [26]. This process is called Cross-spectral Analysis [2]. Proteins with common biological functionality are known to share one significant peak, called the Consensus Frequency [2], which is acknowledged to represent the region responsible for the biological functionality.
Analysis of the Results: Analysis of the result entails identifying the Consensus Frequency (CF) and the utilisation of the amplitude of the Spectral Characteristics of the protein residues at the CF to study their physiological properties. According to the RRM procedure [2], the relationship between the Consensus Frequency (F) and the Peak Position (PP) can be expressed as:
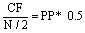 (2)
(2)
This is same as:
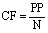 (3)
(3)
where N represents the length of the largest protein in the dataset.
The amplitudes attained by protein residues at the CF demonstrated are useful in determining relationships that exist between the proteins as well as the organisms that harbour them [5,27]. Results can be expressed in terms of percentages depending on the scaling factor. When the scaling function is the maximum value, the highest amplitude obtainable is 1 or 100%.
Method 1: Demonstration of Resonant Recognition Model
Plasmodial peptides P18 and P32 shown in Table 2 have clinically been found to provide immunization against Plasmodium berghei in rodents [28]. They are unequal in length. While peptide P18 has 18 amino acids components, peptide 32 has 32 residues. Using EIIP parameter that is concerned with bio-recognition (specificity of the antibody produced by the peptides) and bio-attachment (relating to the sensitivity of the antibody), RRM procedure is first demonstrated and then used to calculate the degree of bio-recognition and binding interaction. Potency of a vaccine is determined by its specificity and sensitivity. In this study, Plasmodial peptides P18 and P32 are used to demonstrate the RRM procedures.
Demonstration Step 1: Conversion of the Protein Residues into Numerical Values of EIIP Parameter: The entire RRM process is demonstrated in Tables 3 and 4.
| S/No | Seq (P32) | EIIP | CS | CF | Seq (P18) | EIIP | SC | CF |
|---|---|---|---|---|---|---|---|---|
| 1 | I | 0.0000 | - | E | 0.0058 | - | - | |
| 2 | E | 0.0058 | 0.6325 | W | 0.0548 | 0.4778 | 0.3022 | |
| 3 | Q | 0.0761 | 0.4019 | S | 0.0829 | 0.6211 | 0.2496 | |
| 4 | Y | 0.0516 | 0.1830 | P | 0.0198 | 0.5836 | 0.1068 | |
| 5 | L | 0.0000 | 0.6461 | C | 0.0829 | 0.4260 | 0.2752 | |
| 6 | K | 0.0371 | 0.3963 | S | 0.0829 | 0.5025 | 0.1992 | |
| 7 | K | 0.0371 | 0.3891 | V | 0.0057 | 0.6171 | 0.2401 | |
| 8 | I | 0.0000 | 0.2667 | T | 0.0941 | 0.1758 | 0.0469 | |
| 9 | K | 0.0371 | 0.4000 | C | 0.0829 | 0.0920 | 0.0368 | |
| 10 | N | 0.1263 | 0.2810 | G | 0.0050 | 0.5965 | 0.1676 | |
| 11 | S | 0.0829 | 0.7978 | N | 9.1263 | 0.6975 | 0.5564 | |
| 12 | I | 0.0000 | 0.5506 | G | 0.0050 | 0.7390 | 0.4069 | |
| 13 | S | 0.0829 | 1.0000 | I | 0.0000 | 1.0000 | 1.0000 | |
| 14 | T | 0.0941 | 0.3502 | Q | 0.0761 | 0.4411 | 0.1545 | |
| 15 | E | 0.0058 | 0.3770 | V | 0.0057 | 0.5383 | 0.2029 | |
| 16 | W | 0.0548 | 0.7558 | R | 0.0959 | 0.5850 | 0.4422 |
Table 3: Summary of the RRM Procedure: P16 and P32.
| S/No | Seq (P32) | EIIP | CS | CF | Seq (P18) | EIIP | SC | CF |
|---|---|---|---|---|---|---|---|---|
| 17 | S | 0.0829 | 0.7558 | I | 0.0000 | 0.5850 | 0.4422 | |
| 18 | P | 0.0198 | 0.3770 | K | 0.0371 | 0.5383 | 0.2029 | |
| 19 | C | 0.0829 | 0.3502 | - | 0 | 0.4411 | 0.1545 | |
| 20 | S | 0.0829 | 1.0000 | - | 0 | 1.0000 | 1.0000 | |
| 21 | V | 0.0057 | 0.5506 | - | 0 | 0.7390 | 0.4069 | |
| 22 | T | 0.0941 | 0.7978 | - | 0 | 0.6975 | 0.5564 | |
| 23 | C | 0.0829 | 0.2810 | - | 0 | 0.5965 | 0.1676 | |
| 24 | G | 0.0050 | 0.4000 | - | 0 | 0.0920 | 0.0368 | |
| 25 | N | 0.1263 | 0.2667 | - | 0 | 0.1758 | 0.0469 | |
| 26 | G | 0.0050 | 0.3891 | - | 0 | 0.6171 | 0.2401 | |
| 27 | I | 0.0000 | 0.3963 | - | 0 | 0.5025 | 0.1992 | |
| 28 | Q | 0.0761 | 0.6461 | - | 0 | 0.4260 | 0.2752 | |
| 29 | V | 0.0057 | 0.1830 | - | 0 | 0.5836 | 0.1068 | |
| 30 | R | 0.0959 | 0.4019 | - | 0 | 0.6211 | 0.2496 | |
| 31 | I | 0.0000 | 0.6325 | - | 0 | 0.4778 | 0.3022 | |
| 32 | K | 0.0371 | - | - | 0 | - | - |
Table 4: Summary of the RRM Procedure: P16 and P32 (continue).
The Alphabetic codes in the sequences of the peptides P18 and P32 demonstrated in Table 2 are interchanged with the corresponding EIIP values shown in Table 1 in order to obtain their numerical sequences (signals) as displayed in Figure 1. In this numerical form, the peptides can be analyzed using Fourier Transform.
Demonstration Step 2: Zero-padding/Up-sampling: As noted in Table 2, peptide P18 is shorter than the P32 by 14 amino acids compositions. Therefore, 14 zeros are added to P18 so as to bring them to same window length of 32. This is demonstrated in Figure 3. Plots of these numerical sequences (without padding) are shown in Figure 2.
Demonstration Step 3: Fast Fourier Transform (FFT): The numerical signals of Peptides P18 and P32 shown in Figure 3 are then decomposed and processed by means of Fast Fourier Transform (FFT), a faster algorithm for the Discrete Fourier Transform (DFT) using equation:
y = fft(x) (4)
The outcome of the Fourier Transform decomposition yields 32 sets of imaginary, real and absolute values called Spectral Characteristics (SC), which graphically describes the bio-recognition and bio-adhesion of the protein residues/peptides. The plot of SC presents a symmetric (mirror) image [29]. As a result, half the values are considered and the x-axis is scaled to 0.5. The zero frequency of the spectrum also termed” DC component” which is the average value of the signal [1] is discarded. Consequently, 15 sets of values are obtained from the decomposition of the 32 protein residues. The Spectral Characteristics (SC) and Cross Spectral (CS) features of the P18 and P32 are shown in Figure 5.
Demonstration Step 4: Cross-Spectral Analysis: Cross-Spectral analysis represents the point-wise multiplication of the Spectral Characteristics. The length of the largest peptide P32 is 32. Tables 3 and 4 are the demonstration of the entire RRM procedure on the P18 and P32. As shown in Table 4, there are no protein residues at positions 19 to 32 of the P18 and as such, they are zero-padded. The sequences are then translated using EIIP, processed by means DFT to yield Spectral Characteristics (SC) and point-wise multiplied to generate the Cross Spectral (CS) features.
As demonstrated in Table 3, the point-wise multiplication at position 2 of the Spectral Characteristics of P32 (0.6325) and P18 (0.4778) yields 0.3022. This process is carried out all through the sequences. The plot of the CS values yields a symmetric image Figure 4 which is halved and scaled with 0.5. Also,”DC component” of the CS is discarded. Therefore, out of the 32 sequences, 15 values of the Cross Spectral (CS) features are also obtained and illustrated in Figure 5. The plot of the Spectral Characteristics of P18 and P32 are shown in Figure 6 while their Cross-Spectral Characteristics is displayed in Figure 7
Consensus Frequency: According to the RRM procedure [2], the relationship between the Consensus Frequency (F) and the Peak Position (PP) can be expressed as:
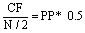 (5)
(5)
This is same as:
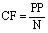 (6)
(6)
where N represents the length of the largest protein in the dataset.
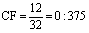 (7)
(7)
Based on equation 7, the CF of P18 and P32 is 0.375.
The Consensus Frequency for the two the Plasmodial peptides P18 and P32 is 0.375 (Position 12).
Using the amplitude value of the proteins and peptides at the CF, biological relationships between the proteins analysed can be identified and their biological functionalities calculated.
Method 2: Information Spectrum Method (ISM)
Information Spectrum Method (ISM), like the RRM is a Digital Signal Processing-based technique that considers protein primary structures or physiological functionalities amino acids sequences of protein represented by numbers using, unlike the RRM, any of the Amino Acids Parameter [2,6,30]. Graphic representation of the biological characteristics by means ISM is referred to as the Informational Spectrum (IS) [2]. Furthermore, point-wise multiplication of the Spectral Characteristics results which reveals shared biological functionalities by proteins demonstrated as a prominent peak called Consensus Frequency (CF) is referred to as Common Informational Spectrum (CIS) [2]. ISM procedure has been used to investigate principal arrangement in Calcium binding protein [30], Influenza viruses [6].
Information Spectrum Method (ISM) uses the same procedure as the RRM. It involves three main steps. Like the RRM, the steps include the conversion the alphabetic code of amino acids sequences into numerical values using amino acids scale that relates to the interaction under investigation. This is followed by the processing of the numerical sequences (signals) using discrete Fourier Transform (DFT). Absolute values of the complex DFT represented as a plot called Informational Spectrum (IS) discloses the information embedded in the protein residues. The y-axis (Amplitude) signifies the contribution in terms of susceptibility or resistance by each sequence while the x-axis (Frequency) determines the position of biological interaction. The third step entails obtaining Common Informational Spectrum (CIS). CIS compares the activities of proteins, which have common biological functions. CIS is the point-wise multiplication of Informational Spectrum (IS).
Demonstration of Informational Spectrum: ISM and the RRM procedure are same. Like in the case of RRM, ISM procedure starts with the conversion of the alphabetic codes of the P18 and P32 peptide residues as shown in Figure 2 into numerical sequences using CHAM830107 parameter in Table 5. The CHAM830107 translated sequences obtained thereafter are then processed by means of Discrete Fourier Transform to obtain Informational Spectra (IS) of P18 and P32.
| Amino Acid | Three Letter Code | One Letter Code | CHAM830107 |
|---|---|---|---|
| Alanine | Ala | A | 0.00 |
| Arginine | Arg | R | 0.00 |
| Asparagine | Asn | N | 1.00 |
| Aspartic acid | Asp | D | 1.00 |
| Cysteine | Cys | C | 0.00 |
| Glutamine | Gln | Q | 0.00 |
| Glutamic acid | Glu | E | 1.00 |
| Glycine | Gly | G | 1.00 |
| Histidine | His | H | 0.00 |
| Isoleucine | Iso | I | 0.00 |
| Leucine | Leu | L | 0.00 |
| Lysine | Lys | K | 0.00 |
| Methionine | Met | M | 0.00 |
| Phenylalanine | Phe | F | 0.00 |
| Proline | Pro | P | 0.00 |
| Serine | Ser | S | 0.00 |
| Threonine | Thr | T | 0.00 |
| Tryptophan | Trp | W | 0.00 |
| Tyrosine | Tyr | Y | 0.00 |
| Valine | Val | V | 0.00 |
Table 5: A parameter of charge transfer capability (CHAM830107).
Using equation 6, the Consensus Frequency is recognized as:
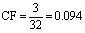 (8)
(8)
Therefore, the CF of the two Plasmodial peptides based on CHAM830107 is 0.094.
Based on the amplitudes of the protein residues at the CF identified, further investigations into the biological behaviour of the proteins can be disclosed by means of the ISM and amino acids parameter, CHAM830107 engaged.
Materials
Amino acids sequences of the Plasmodial Circumsporozoite, HIV glycoprotein41 from HXB2 isolate are retrieved from UNIPROT [31]. P18, P32, and Enfuvirtide and Sifurvide are obtained from the literature. While P18 and P32 amino acids sequences are recognized as being studied as starter materials for vaccine development [28], the Enfuvirtide amino acids sequence has been examined along with that of the Sifuvirtide, which is noted to be a bio-medically engineered peptide, obtained from the HIV gp41 C-terminal of the Heptad Repeat (CHR) sequence of the HIV subtype E [13,32].
Preliminary Clinical Studies: P18 and P32
The two sets of pharmacologically active components are the Plasmodial peptides P18 and P32, as well as anti-HIV Fusion peptides namely; Enfuvirtide (T20) and Sifuvirtide are first clinically studied. Using the related Amino Acids Parameters, these pharmacologically activities are calculated by means of Digital Signal Processing technique and correlated with our computational outcomes.
Peptides P18 and P32 from the Plasmodium falciparum have been identified to inhibit Plasmodium berghei invasion of Hep-G2 while P32 is found to protect immunised mice [28]. Interactions between the CS and the hepatocytes and subsequent invasion of the Hep-G2 by the Plasmodial sporozoites have been recognised [33,34]. Prior to interaction, the two proteins must bio-recognise and bind. This interaction is governed by EIIP parameter [35]. Negatively charged carboxyl group of the GAGs have been found to partake in the binding of the CS to HSGP [26]. As a result, amino acids parameters that engage charges (Positive and Negative) are employed in this study.
Results and Discussions: P18 and P32
In this study, EIIP (Table 2) and five (5) other amino acids parameters such as the amino acids parameter with descriptor CHAM830107 (Table 5), which are based on charges including CHAM830108, FAUJ880111, FAUJ880112 and KLEP840101 are retrieved from [16] and engaged in the demonstration of the ISM analysis of the P18 and P32 peptides sequences shown in Table 2 and further used to calculate their biological functionalities. The results are presented in Table 6.
| Sequence | EIIP | CHAM830107 | CHAM830108 | FAUJ880111 | FAUJ880112 | KLEP840101 |
|---|---|---|---|---|---|---|
| P16 | 1.000 | 1.000 | 1.000 | 1.000 | 0.716 | 0.888 |
| P32 | 1.000 | 0.898 | 0.811 | 1.000 | 1.000 | 1.000 |
| CF (position) | 0.375(12) | 0.094(3) | 0.438(14) | 0.032(1) | 0.156(5) | 0.094(3) |
| CF (Amplitude) | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
Table 6: Calculated Biological Functionalities of P18 and P32.
As shown in Figures 6 and 7, both P18 and P32 have maximum amplitudes of 1.00 at the CF, which is at Position 12. This appears to indicate 100% bio-recognition and bio-attachment for the two peptides (P18 and P32). This characteristic determines specificity of interaction between the peptides and the ligands that produce the immunizing antibodies, a property that determines vaccine potency.
It is therefore demonstrated in this study that both P18 and P32 have amplitude of 1.00, which suggest 100% affinity for the target protein that will elicit the production of the antibody necessary for the immunization against Malaria. These are peptides that are still being studied for possible engagement in the designing of vaccines. Vaccines are required to demonstrate specificity in action so that they could produce specific antibody that would neutralise specific antigen. The outcome of this study appears to suggest that both peptides are suitable starter materials for the design of Malaria vaccines by their specificity (precision in the production of Malaria vaccine rather than any other disease) as demonstrated in their ability to bio-identify and bio-attach to the target protein.
Though the results of the EIIP (Figure 6) and FAUJ880111 (Table 6) parameters demonstrated equi-potency by maintaining same maximum (100%) interaction (Figure 2 and Table 6), other parameters disclose unequal potencies. Using CHAM830107 and CHAM830108, our results reveal higher and maximum (100%) interaction for the P18 (Table 6). For the P32, the CHAM830107 parameter yields 89.8% interaction while the CHAM830108 produces 81.1% interaction.
On the other hand, the FAUJ880112 and KLEP840101-based analyses show higher and maximum (100%) for the P32. These seem to indicate higher sensitivity; hence potency for the P32 peptides in terms of the FAUJ880112 and KLEP840101 parameters. As shown in Table 6, FAUJ880112 parameter revealed 100% interaction for the P32 at the CF and 71.6% for the P18. In the same manner, KLEP840101- based analysis reveals 100% interaction for the P32 and 88.8% for the P18. These appear to suggest different sensitivity and as such, different capability for the antibody generated in immunizing host organisms for Malaria except for the EIIP and FAUJ880111 which demonstrated equi-potency.
The six amino acids parameters studied and shown in Table 6 therefore express the specificity (bio-recognition and bio-attachment to the target proteins that direct the production of antibody for Malaria only). In the case of EIIP parameter which measures specificity, the level of protection from the Malaria (potency) by the two Plasmodial peptides used (P16 and P32) is the same while the degree of sensitivity as provided by the other parameters vary. These physiologic indices (amino acids parameters) describe the therapeutic value of the vaccines produced. In both peptides, it is disclosed all interactions by means of the amino acids parameter engaged are above 50%, which appears to suggest that both could be suitably used in the on-going study for the design of anti-Malaria vaccine.
Preliminary Clinical Studies: HIV Fusion Inhibitors
Sifuvirtide, a product of Biomedical engineering is claimed to be more potent, highly effective against Enfirvitude resistant strains, safer and better tolerated than the Enfirvitude also called T20 [32]. Prototypic Peptide 7 has been obtained from gp41 of HIV-1 subtype E [37,38]. By means of Biomedical engineering approaches, it was redesign to obtain Sifuvirtide through the introduction of the salt bridge. This led to increased helicity, stability of the Six Helix Bundle (coiledcoil) formed by the anti-HIV/AIDS peptides and the target protein, N-terminus Heptad Repeat (NHR). This determines the anti-HIV/ AIDS potency [37,38].
Biomedical engineering design of the Sifuvirtide was achieved by the introduction of the charged amino acids namely, glutamic acid and lysine. Also to provide hydrophobicity pocket, Glutamic acid (E) at position 119 (shown in Table 7 as red, bold and elongated alphabetic code) was replaced with Threonine (T) [32]. Furthermore, Serine was added to the N-terminus so as to increase its stability [32].
| Sifuvirtide Prototypic Peptide(117-151) | WIEWEREISNYTNQIYEILTESQNQQDRNEKDLLE |
Table 7: Prototypic Peptide of Sifuvirtide.
As demonstrated above, Biomedical engineering of thePrototypic Peptide 7 into the Sifurvitude altered the helicity and hydrophobicity properties and as such, three Alpha Helix-related amino acids parameters namely, BURA740101, PONP800104, and PRAM900102 are engaged in this study. Furthermore, five Hydrophobicity-based Amino Acids Parameters including ARGP820101, ENGD860101, FASG890101, JURD980101 and WOLR790101 are employed in the calculation of the pharmacological activity of the two Fusion inhibitors (anti-HIV/AIDS agents). The target protein, N-terminus Heptad Repeat (NHR) is also investigated to show its contribution in interaction. The outcomes of the bio-recognition, bio-affinity and other interactions computationally investigations are as shown in Table 8 using .9 parameters.
| S/No | Parameter | Enfuvirtide | Sifuvirtide | NHR | CF |
|---|---|---|---|---|---|
| 1 | EIIP | 0.815 | 1.000 | 0.778 | 0.281(9) |
| 2 | BURA740101 | 0.467 | 0.639 | 0.479 | 0.562(18) |
| 3 | PONP800104 | 0.745 | 0.764 | 1.000 | 0.283(13) |
| 4 | PRAM900102 | 0.648 | 0.785 | 0.821 | 0.283(13) |
| 5 | ARGP820101 | 0.810 | 1.000 | 0.915 | 0.283(13) |
| 6 | ENGD860101 | 0.637 | 0.851 | 0.907 | 0.283(13) |
| 7 | FASG890101 | 0.841 | 0.885 | 1.000 | 0.283(13) |
| 8 | JURD980101 | 0.828 | 0.343 | 0.237 | 0.283(13) |
| 9 | WOLR790101 | 0.858 | 0.290 | 0.208 | 0.283(13) |
Table 8: Calculated Biological Functionalities of Enfuvirtide, Sifuvirtide and NHR.
Results and Discussions: HIV Fusion Inhibitors
The outcome of the Common Informational Spectrum (CIS) of the Enfuvirtide, Sifuvirtide and NHR shown in Table 8 using nine Amino Acids Parameters reveals that the CF is at 0.283(13) for all as exemplified by the hydrophobicity-based parameter PONP800104 and Alpha Helix-base ARGP820101 Figure 10 except for the EIIP which is at 0.281(9), and BURA740101 at 0.562(18) though another peak is noticed at position 9 (Figure 11).
From the nine parameters used and shown in Table 8, the amplitude values for the Spectral Characteristics of the Sifuvirtide (using EIIP) is 1.00 (Table 8), which appears to suggest 100% affinity while that of the Enfuvirtide, is 0.815 (Table 8) suggesting 81.5% affinity.
Except for Amino Acids Parameter with descriptor name WOLR79010 (Table 8), Sifuvirtide demonstrated higher amplitude values hence higher biological functionality than the Enfuvirtide. However, based on the nine parameters engaged, the average percentage (%) calculated Biological Functionalities in the Enfurvitide (72.33%) appear to be less than that obtained in the Sifurtivide (85.42%) as shown in Table 8. This appears suggest that Sifurvide may possess higher therapeutic value as claimed in the preliminary clinical studies. This study is based on the amino acids parameters engaged which are considered to determine the mechanism of actions of the two Fusion Inhibitors of the HIV. Clinically, Sifuvirtide has been suggested to exhibit better efficacy than the Enfuvirtide [32]. Such clinical findings include that the fact that it has six fold higher HIV fusion inhibitory activity. It has been shown that Sifuvirtide not only forms Six Helix bundle (SHB) with target protein (NHR) but blocks other peptides. This is unlike Enfuvirtide.
These claims have preliminarily authenticated by means of CD spectroscopy, a clinical approach. CD spectroscopy has indicated 93% alpha helical content for Sifuvirtide while the Enfuvirtide has none [32]. These factors may have been responsible for the claim that Sifuvirtide is more efficacious as it provides more interaction (average of 85.42%, as shown in Table 8) with the NHR (Table 8), which is known to possess hydrophobic pockets. As noted in Table 8, NHR which interacts with the two anti-HIV/AIDS drugs (Enfuvirtide and Sifuvirtide) offers high level of interaction (87.07%) to these antiretroviral agents.
These findings appear to agree with the calculated Biological functionalities carried out in this study using 9 amino acids parameters that relate to the physiologic indices employed in the clinical experiments (Helicity and Hydrophobicity).
Calculating Biological functionalities by means of Digital Signal Processing techniques appears to be beneficial in the design and development of drugs and vaccines. The introduction of Reverse Vaccinology in the designing of vaccines has resulted in the use of protein fragments or peptides as starter materials for vaccine design. Calculating the Biological characteristics of these peptides rather than obtaining results through clinical experimentation remains a more rational approach. Also, comparisons of Pharmacological activities of the peptide-based drugs by means of calculating their Biological functionalities are a faster and resource saving approach.
Two Digital Signal Processing techniques namely Resonant Recognition Model (RRM) and Informational Spectrum Model (ISM) are employed in calculating the Biological behaviours of two peptides (P18 and P32) which are being investigated for possible incorporation into the materials for the designing of anti-Malaria vaccines. Also, the Biological features of two Fusion inhibitors known as Enfuvirtide (T20) and Sifuvirtide are calculated and the outcomes compared with initial clinical findings. Furthermore, studying of the effect of mutations on the Biological functionalities by delivering them numerically also quickens the application of the outcomes.
Our results revealed that both P18 and P32 share maximum affinity (100%) which seems to suggest that they offer high specificity that could result in the production of an appropriate antibody. Other interactions studied by means of amino acids parameters engaged which relate potency (sensitivity and neutralization power) of the antibody produced are also found to be high.
Furthermore, our findings disclose that Sifuvirtide (0.8542) has higher average amplitude, which suggest higher interaction (85.42%) than the Enfuvirtide with average amplitude of 0.7233 suggesting 72.33% interaction. This is based on the 9 amino acids parameters which are associated with the physiologic indices clinically, examined indicating higher interaction. This therefore suggests that Sifuvirtide may be more efficacious based on the 9 amino acids parameters studied. This result is found to correlate with clinically derived outcome. However, these results are not interpreted in terms of Pharmacokinetics and Pharmaco-dynamic activities like solubility, absorption, shelf-live, toxicity, distribution, excretion, etc. As a result, our findings do not suggest acceptability of the Sifuvirtide for the HIV/AIDS management.
This study therefore appears to demonstrate that calculating Biological functionalities is an easier approach to comparing Pharmacological activities of drugs. It also helps determine the Biological activities of peptide components of drugs and vaccines. Manipulation of the amino acids sequences for optimal Biological activities can only be simplified when their Biological functionalities are known and better, delivered or presented in numerical terms.