Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research Article - (2021)Volume 9, Issue 2
Probiotics may represent an appropriate alternative to oral therapy by specifically targeting pathogens during the successive phases of biofilm “formation”. The aim of this work was to find ways to complement and strengthen this anti-biofilm action through examining the effects of 13 probiotics on the “preformed” three-dimensional biofilm structure. An in vitro dental multi-species biofilm model has been developed using pioneer-colonizing streptococci strains Streptococcus mutans, Streptococcus sobrinus, and Streptococcus oralis subsp. oralis with the addition of Actinomyces naeslundii, to obtain highly heterogeneous strong biofilm architecture. Among the different probiotic genera tested, only the genus Bacillus had a significant disruptive impact on biofilm. B. subtilis NOH (Natto Original Habitat) isolated from a traditional Japanese fermented food (natto) was the most effective disrupter, able to destroy 39% of pre-formed biofilm. Some results have suggested the presence of biosurfactants in its supernatant. Unlike B. subtilis NOH, the B. subtilis CU1 (CNCM I-2745) strain showed strong antibacterial properties against pathogenic organisms and blocked biofilm development at high probiotic concentration. A dual strategy of applying agents with disruptive (B. subtilis NOH) and antimicrobial (B. subtilis CU1) activity could be an interesting approach, with disassembling of the matrix structure exposing pathogenic organisms to more efficient killing.
Biofilm; Probiotics; Oral health; Bacillus subtilis
Oral health is a major concern, given the high prevalence of cavities, gingivitis, halitosis, and periodontitis worldwide. According to the World Health Organization, more than 60% of children and almost 100% of adults experience caries, and the financial cost of oral care is estimated at 5% of the health budgets of developed countries [1]. The oral cavity is home to around 700 species of microorganisms, among which both beneficial and pathogenic microorganisms capable of adhering and forming biofilms, can coexist and organized themselves into a consortium of bacteria, viruses, and fungi [2].
Oral diseases originate from a dysbiosis of this oral community. Healthy people usually maintain a balance between both groups of microorganisms, but factors such as stress, diminished immunity, or a high-sugar diet can induce a shift in these populations toward the pathogenic group [2,3]. The proportion of pathogenic strains then increases within the biofilm, with important consequences for oral health. Some pathogens, such as Streptococcus mutans and Streptococcus sobrinus, acidify the oral environment through lactic acid production from sugar metabolism, promoting caries development [4]. In addition, the formation of a pathogenic biofilm in the periodontal pocket can result in gingivitis and periodontitis [5].
Biofilm consists of a matrix of soluble and insoluble polysaccharides (EPS), which confer protection on and provide energy for the bacteria [6]. These sugars are mainly glucans, produced by the action of glucosyltransferases (GTFs) using sucrose as a substrate [7]. S. mutans produces several GTFs, including GTF-B and GTF-C, which are responsible for extracellular polysaccharide that consists of α-(1,3)-linked glucose residues in main chains and α-(1,6) bonds in side chains [7]. S. sobrinus along with Streptococcus oralis, subsp. oralis an early colonizer of the mouth, also produces GTFs, contributing to the development of the biofilm matrix [8-11].
In addition to soluble and insoluble glucans, biofilm contains proteins, nucleic acids, lipids, and other biopolymers [12,13]. The matrix they form has a protective function against environmental conditions and antibiotics. The spatial proximity of the bacteria is ideal for promoting exchange of resistance genes, enhancing the overall resistance and pathogenicity of the biofilm [14,15]. Furthermore, the biofilm colonizes every surface of the oral cavity, from the teeth to periodontal pockets [16]. Given its infectivity, resistance, and widespread presence in the oral cavity, biofilm matrix appears to be an appropriate target for the treatment and prevention of oral diseases [17].
Current oral care products such as mouthwash, antiseptics, and antibiotics are efficient against oral diseases. However, they are indiscriminate and do not specifically target pathogens preferentially over beneficial bacteria, resulting in oral ecosystem imbalance [18]. Moreover, the pathogenic microbes are more effective at recolonizing the oral cavity [19]. Thus, such oral care products may enhance the prevalence of pathogenic strains which contribute to an unbalanced mouth ecosystem that ultimately could leads to more disorders. Consequently, innovative approaches are required to limit oral diseases and restore a more balanced oral community.
Recently, studies have shown that probiotics-microorganisms with a known beneficial impact on some aspects of health-are potential innovative candidates for oral care [20-22]. Indeed, probiotic strains have already been used to improve oral health in the context of diseases such as childhood caries [23,24]. Other studies have shown the potential of probiotics to treat periodontitis and caries, helping to restore homeostasis that cannot be achieved with conventional treatments [25,26].
Different in vitro model systems have been developed to study complex biofilms. For the screening of anti-biofilm agents, the system should be easy to use, have high reproducibility and high efficiency. The simplest model is a closed culture system, on different surfaces for the growth of oral biofilm, such as multiwell plates [27]. Single-species biofilm in vitro model cannot represent the interaction and complex functions of microorganisms associated with oral multispecies biofilms [28]. For example, the expression of gtfB and gtfC genes is enhanced in the presence of other oral bacteria like Actinomyces naeslundii and Streptococcus oralis [28,29].
In this study, four microorganisms were selected for the development of the biofilm, based on previous work [30]. An in vitro dental plaque model, that contain microorganisms associated with healthy and pathogenic enamel biofilms, has been developed using Streptococcus mutans, S. sobrinus and S. oralis subsp. oralis with the addition of Actinomyces naeslundii, a significant member of the initial colonizers of tooth surfaces and dental biofilm formation [29].
The aim of this work was to relate the 96-well microtiter plate static biofilm system with a relevant multispecies dental caries model that could be reproducibly grown to allow the identification of probiotic anti-biofilm therapies. Our long-term objective was to develop a new probiotics-based product for oral care. As a step toward this goal, we evaluated the anti-biofilm potential of 13 probiotic strains, specifically focusing on their ability to disrupt a pre-formed biofilm. Depending on the target, it is important to choose the appropriate quantification method. So, if disruption of preexisting biofilm is the objective, methods which quantify total biomass of the biofilm should be applied [31].
Staining of biofilms grown in microtiter plates is extensively used by researchers to screen and compare biofilm formation and disruption. Among the methods described in the literature, Crystal Violet (CV) staining is the most frequently used for biofilm biomass quantification [32]. This basic dye binds negatively charged molecules and thus stains both bacteria and the surrounding biofilm matrix. Safranin staining provided similar results as crystal violet, but with higher reproducibility and less toxicity [32].
During this work, the total biofilm biomass of the developed model, determined by safranin dye, is the essential parameter characterized, as this provides insight into the structural stability of the formed biofilm. It is more frequently used for assessing the sensitivity of a treatment against the extracellular matrix of the biofilm, rather than the measurement of microbial populations and their interactions.
Thus, the safranin staining intensity, measured by spectrophotometry, will allow quantifying both the adhesion of the biofilm and its biomass.
Bacterial strains and growth conditions
Biofilm bacterial strains: Streptococcus mutans DSM 20523 serological group c (ATCC 25175), isolated from carious dentine, Actinomyces naeslundii DSM 43013 (ATCC 12104) isolated from human sinus, Streptococcus oralis subsp oralis DSM 20627 (ATCC 35037) known as gtfR-positive isolated from human mouth and Streptococcus sobrinus DSM 20742 (ATCC 33478) isolated from human dental plaque were used to generate mixed-species biofilms [11].
Pre-cultures of all bacterial strains used for biofilm formation were cultured on Brain Heart Infusion (BHI) supplemented with 2.5 g of peptone from casein digest, 5 g of yeast extract, and 5 g of glucose (Bio-Rad® BHI 217) at 37°C under microaerophilic conditions for 24 h.
Probiotic strains: Probiotic strains used in this study are listed in Table 1 and come from various sources including commercially available lozenges. They were cultivated on the supplemented Brain Heart Infusion medium (Bio-Rad® BHI 217). All strains were cultivated at 37°C under aerobic or anaerobic conditions, as warranted.
| Name | Strain origin | Probiotic mechanisms of action | References |
|---|---|---|---|
| Bacillus coagulans | DSMZ 1 | Reduction in cariogenic microorganisms | [33] |
| Bacillus subtilis | CNCM I-2745 (CU1) |
Antimicrobial lipopeptide production | [34] |
| Bacillus subtilis NOH | Natto Food | View results data | - |
| Bacillus subtilis var. natto | DSMZ 402 | Proteolytic enzymes production | [35] |
| Lactobacillus acidophilus | ATCC 4356 | Reduction of Streptococcus mutans adherence, decreased glucan production by S. mutans, and antibacterial activity | [36,21] |
| Lactobacillus gasseri |
Supersmart® | Disruption of mature biofilm formation | [37] |
| Lactobacillus paracasei |
CNCM LA802 | Biosurfactant production and inhibition of S. mutans growth | [38,39] |
| Lactobacillus reuteri | Supersmart® | Reduced expression of genes involved in acid tolerance; quorum sensing (QS) and EPS production | [40] |
| Lactobacillus rhamnosus | ATCC53103 | Reduction of surface tension and emulsifying activity; biosurfactant production | [41] |
| Lactobacillus salivarius | DSMZ 20555 | Peroxide-dependent antimicrobial and anti-biofilm activities, reduced expression of genes involved in acid tolerance, QS and EPS production, growth inhibition of S. mutans, and reduced expression of S. mutans genes gtfB | [40,42] |
| Saccharomyces boulardii |
Enterol | Reduced adhesion and biofilm formation | [43] |
| Streptococcus dentisani |
DSMZ 27089 | Bacteriocin production against S. mutans and S. oralis | [44] |
| Streptococcus salivarius |
K12 | Bacteriocin-like inhibitory substances production | [45] |
Scientific publications on probiotic microorganisms, related oral health claims, and their potential anti-biofilm activities guided our selection of probiotics for this work. The probiotic strains that have been selected can be isolated from many sources such as humans, plants, environment, and foods and belong to a variety of genera including LactoBacillus, Bacillus, Streptococcus, and Saccharomyces. The selected probiotic strains most often reported to exert anti-biofilm activity are listed in Table 1 with some of their anti-biofilm specifications.
Biofilm preparation and anti-biofilm evaluation of probiotics
Our approach to evaluating the ability of a probiotic strain to disturb a performed oral biofilm was based on the Zurich model [46]. Zürich biofilm model is a multi-species model that allows the interspecies associations to be studied with respect to biofilm formation.
Following 24 hours preculture and culture in the relevant medium, most often BHI 217, we measured optical density at 600 nm for each culture. Based on the value, the cultures were mixed at a final dose of 5 × 106 colony-forming units (CFU)/ml in the BHI 217 medium supplemented with 0.25% sucrose. Subsequently, 180 μl of the mix was dispensed onto a sterile 96- well plate, with 8 wells containing medium only as a blank control. The plate was incubated at 37°C under a microaerophilic environment.
After 7 hours of incubation, the medium inside each well was carefully removed and renewed with fresh medium (BHI 217+0.25% sucrose). The plate was again incubated for 17 hours at 37°C, and after a total of 24 hours, the mature biofilm was assumed to be established and ready for testing.
To determine the anti-biofilm effect of the probiotics, we prepared the plate as follows. First, the medium was pipette off and biofilms were washed three times with PBS, following by dispensing of 100 μl of a given probiotic culture at variable inoculum concentrations from 106 to 2 × 108 CFU/ml, into at least six wells for replicate purposes. Untouched wells containing the initial medium and biofilm building strains served as negative controls, whereas the positive controls contained Sodium Dodecyl Sulfate (SDS) 0.5% (w/v) or glucanex 0.125% (Sigma-Aldrich). After 24-hour incubation, quantification was performed as described below.
Biofilm quantification
For biofilm quantification, we used safranin staining [31]. The biofilm was washed twice by dipping the plate into a crystallizer filled with tap water. The water inside each well was then removed via several shakes over a paper towel, followed by the addition to each well of 125 μl of a 0.25% safranin solution (20% ethanol, 80% water) and incubation for 10 minutes at room temperature. The plate was then washed again twice to remove unbound stain. After the plate was dried, we added 125 μl of dimethyl sulfoxide to each well to solubilize the safranin. The amount of biofilm was quantified by spectrophotometry at a wavelength of 492 nm, with medium only as the blank.
B. subtilis NOH supernatant preparation
B. subtilis NOH was cultivated for 24 hours in BHI 217 at 37°C under aerobic conditions. The culture was then centrifuged at 5000 × g for 5 minutes and the supernatant recovered and sterilized by filter-sterilization (Sterile Millex® Filters 0.25 μm).
Surface tension properties of Bacillus NOH culture and supernatant: collapse test
To qualitatively evaluate the tensioactive property of a solution, we used the collapse test, which is based on the “Parafilm M test” described previously [47]. After 100 μl droplets of water were deposited on the surface of a parafilm band, we injected each droplet with 10 μl of the test substance. Using a ruler, we measured the droplet diameter and compared that result to the diameter of a control droplet injected with water. The droplet diameter is expected to be proportional to the degree of spreading, which in turn is proportional to biosurfactant activity or concentration because of decreased surface tension.
The agar diffusion method for antimicrobial screening of Bacillus sp.
To assess the antibacterial activity of Bacillus sp. on biofilm positive strains, we used an agar diffusion method [48]. The tested strain was mixed at a highly concentrated dose (1 × 109 CFU/ml) with a 1.2% agar/water mix at 55°C and poured onto the surface of a Petri dish containing BHI 217 medium. After agar solidification, wells were dug in the agar using a sterile metallic pipe and filled with 100 μl of highly concentrated culture of the tested probiotic. The plate was incubated at 37°C in a microaerophilic environment. After 24 hours, a clear growth inhibition zone around the wells was used to infer that the strain inside the well showed bactericidal action against the tested pathogenic microbes.
Biomass kinetic studies
For the 72 h kinetic studies, planktonic cultures were prepared as described previously in BHI-217, 0.25% sucrose. 125 μl of prepared dilutions with mixed microorganisms (1:1:1:1 of S. sobrinus: S. mutans: S. oralis: A. naeslundii) were plated in each well of 96 well microtiter plates. At each time point in the 72 h study the biofilms were stained with safranin for biomass determination.
Microscope biofilm observation
Biofilm formation in an 8-well chamber slide was performed. It allows us to directly observe a biofilm. The preparation follows the same steps as described above for the 96-well plate. However, the volume is 300 μl instead of 180 μl. Once the experience is finished, the biofilm is washed twice by dipping the plate inside a crystallizer filled with tap water. The water inside each well is then removed by performing several shakes on a paper towel. 150 μl of a safranin 0.25% solution (20% ethanol and 80% water) is added in each well and the slide is incubated for 10 minutes at room temperature. The slide is then washed again twice to remove the unattached stain. After drying, the wells and the joint are removed leaving the biofilm on a regular microscope slide. The biofilm is observed on an optical microscope at a magnification of 1000.
Statistical analysis
After assessing whether data conformed to a normal distribution, One-way Analysis of Variance (ANOVA) followed by test Tukey’s multiple comparisons (α=0.05) was used to investigate significant differences between independent groups of data. A Bonferroni correction was applied to the p value to account for multiple comparisons of the data. Statistical significance was achieved if P<0.05.
Multi-species biofilm formation (model)
The aim of this experiment was to validate a system that can adequately reflect the multi-species interactions in biofilms, while maintaining the simplicity needed to ensure reproducibility and efficiency in screening studies.
Different microbial combinations associated with S. mutans to form dual and multi-species biofilms (three and four strains combination), were achieved by incubating bacterial suspensions (5 × 106 CFU/ml-1) on flat-bottomed 96-well polystyrene microwell plates at 37°C and incubated 24 h. The biomass of the multispecies biofilm is also evaluated with a five to ten reduction of inoculum concentration (1/5 and 1/10 dilutions).
In all cases, as illustrated Figure 1, bacteria adhered directly on polystyrene surfaces. The safranin staining indicated an increase in the biomass for all dual and multi-species models compared to the S. mono-species biofilms. However, we must underline the strong propensity of A. naeslundii to synthesize a biofilm alone. Indeed, each species within the biofilm produced different amounts of biomass when grown in a mono-species biofilm. After 24 hours of growth, the A.n mono-species biofilm and all the multispecies biofilms had a significantly (p<0.05) higher biomass production than the mono-species Streptococci biofilms. A five to ten reduction of inoculum concentration, decreases very slightly, the multi-strains biofilm intensity over 24 hours period. Overall, the four-species produced significantly more biomass than the other S. mutans associated biofilms.
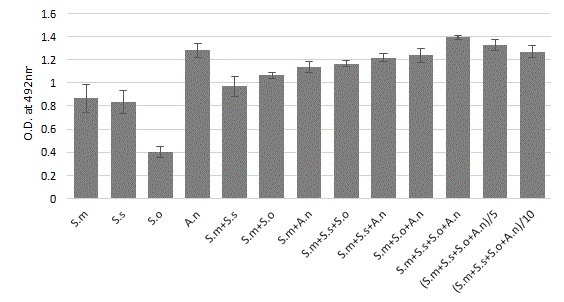
Figure 1: Quantitative spectrophotometric determination (safranin coloration) in biofilm formation of mono, dual and mixed-species culture after 24 h of incubation. Bacterial suspensions were mixed (1:1:1:1 OD600 nm 0.05) to provide an inoculum with a defined final concentration of 5 � 106 CFU/ml. Precultures on BHI of the four individual species were mixed, diluted to a final OD600=0.05; 0.01 and 0.005 and used for inoculation of the plates. Streptococcus mutans (S.m), sobrinus (S.s), oralis (S.o) and Actinomyces naeslundii (A.n). The mean ± standard deviation for 12 replicates is illustrated.
In order to study the influence of anti-biofilm agents on the extracellular matrix of a preformed biofilm, it is necessary to identify, the different stages of its development over time (growth, maturation) by monitoring the kinetic of biomass in our model conditions.
Kinetic of 72 h biofilm process
The BHI sucrose 0.25% media supported the viability and biomass production of multispecies biofilms. Kinetic studies over 72 h, from the moment the strains are seeded, demonstrated a stable biofilm period between 24 and 72 h, after a 24-hour building phase (Figure 2).
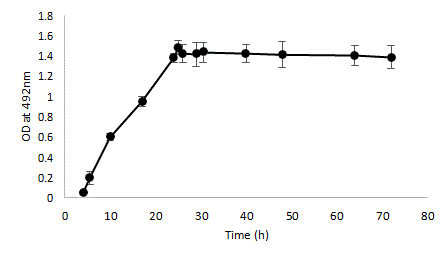
Figure 2: Kinetic of multi-strains biofilm development by quantitative spectrophotometric determination (safranin coloration) of biofilm biomass of a mixed-species culture. Bacterial suspensions were mixed (1:1:1:1) to provide an inoculum with a defined final concentration of 5 Ã? 106 CFU/ml. Precultures on BHI of the four individual species (A. naeslundii, S. mutans, S. oralis and S. sobrinus) were mixed, diluted to a final OD600=0.05 and used for inoculation of the plates. Error bars represent the standard deviations of the means of six experimental replicates.
The four-species mixed biofilm model biomass was stable across all the tested inoculum concentrations and represents a good model for the screening of potential disrupting agents. The main advantages of the method are its high reproducibility and repeatability and rapid analysis of biofilm biomass.
Impact of probiotic monostrain cultures on 24 hpreformed biofilm
In our in vitro model, the capacity of probiotics to disperse a 24 h preformed pathogenic biofilm constructed with the reference microbes (Actinomyces naeslundii, Streptococcus mutans, Streptococcus oralis, and Streptococcus sobrinus) was determined by safranin staining and biofilm total biomass quantified by spectrophotometry. So, specific disrupting capacity of a preformed biofilm was expected to be inversely proportional to the optical density at 492 nm.
Established biofilms were treated with probiotic culture at high inoculum (108 CFU/ml), matrix biofilm breakdown was evaluated, and results are depicted in Figure 3.

Figure 3: Effect of probiotic bacteria on pre-formed biofilm. Optical density (safranin coloration) of A. naeslundii, S. mutans, S. oralis and S. sobrinus biofilm. Data are expressed as the mean ± standard deviation of five to ten independent experiments in sextuplicate (big sample size, at least 30). **p<0.01, ***p<0,001 significance was determined by comparison to the untreated control. Reference positive control: SDS 0.5%.
Among the probiotics tested, only genus Bacillus shows disturbing activities on the preformed biofilm, despite the probiotic high inoculum doses used. Bacillus subtilis NOH isolated from Natto food and Bacillus subtilis CUI strain reduced significantly (p<0.001) the total biofilm biomass. The mature biofilm reduction was assessed by safranin optical density drop between the control and the probiotic suspension, from 1.44 (control) to 0.88, 1.12 and 1.29 corresponding to a biofilm destruction of 39% for B. subtilis NOH, 22% for B. subtilis CU1, and only 10% for B. subtilis var. natto.
On the other hand, most strains of LactoBacillus (L. acidophilus, L. paracasei, L. reuteri, and L. salivarius) induced sometimes a significant increase (p<0,001) of up to 11% in biofilm intensity. The other strains (L. rhamnosus GG, S. boulardii, S. salivarius K12, S. dentisani, and L. gasseri) had no significant impact on the mature biofilm.
Pre-formed biofilm disruption: Bacillus subtilis NOH cell density
To determine the ability of Bacillus NOH to disrupt pre-formed biofilms, the test biofilms were formed in microtiter plate wells for 24 h. The pre-formed biofilms were then treated with different Bacillus NOH culture dilutions with fresh medium to reached final cell concentration from 106 to 2.108 CFU/ml. The residual biofilm was estimated 24 h later using the safranin assay. Microtiter plate wells containing biofilm with medium without Bacillus strain were used as controls during the experimentation. The data related to these experiments are depicted as the average values of six observations and error bars indicate the standard deviation (Figure 4).
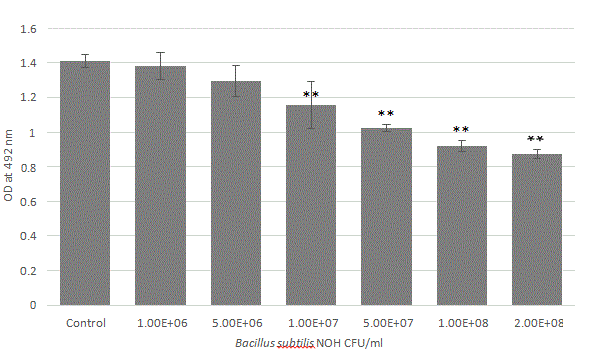
Figure 4: Bacillus subtilis NOH cell density effect on pre-formed biofilm disruption. Biofilms were grown for 24 h, and then treated for 24 h with Bacillus at different concentration (CFU/ ml), and changes in biofilm biomass were evaluated. For the biofilm biomass, the results were measured as the optical density of the safranin staining at 492 nm. All trials were carried out in triplicate on three independent experiences. Data represents mean ± standard deviation, statistical analysis of treatments was compared to the untreated control (*p<0.05, **p<0.01).
This experiment shows that Bacillus subtilis NOH disrupted biofilm biomass in a cell density dependent manner. Significant biomass reduction almost requires 5 × 106 CFU/mL (p<0.01). Nevertheless, probiotic population above 108 cells/ml is recommended to achieve notifiable biofilm destruction near 40%.
Kinetic dispersion of Bacillus NOH on 24 h-preformed biofilm
To assess the capacity of Bacillus NOH on a preformed 24 h biofilm, it is interesting to compare the normal evolution of this preformed biofilm (control) compared to its evolution in the presence of the Bacillus strain. Figure 5 shows that only 1 hour is needed to start biofilm destruction in presence of B. NOH while the biofilm is stable in the absence of the strain.
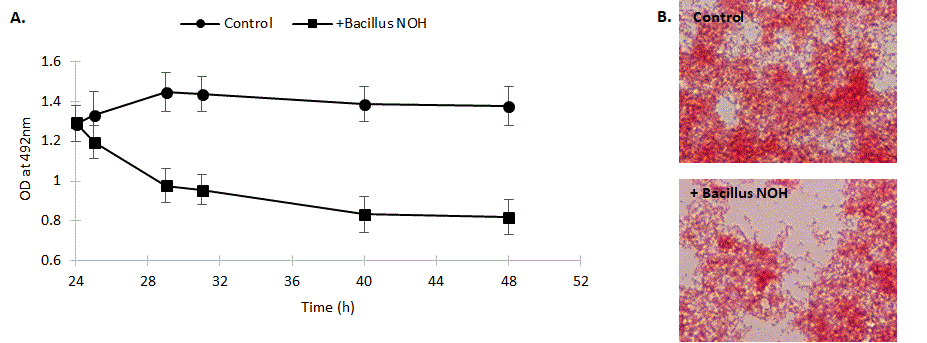
Figure 5: Effect of Bacillus NOH (versus control) on the destruction kinetic of a pre-formed multi-species biofilm with A. naeslundii, S. mutans, S. oralis and S. sobrinus. A. Determination of biomass after safranin staining by optical density at 492 nm. Data are expressed as the mean ± standard deviation of five independent experiments. B. Light microscopic images represent the biofilms 24 h after treatment with B. subtilis NOH versus non-treated control.
To visualize the ability of Bacillus to degrade mature biofilm, 24 h-preformed biofilms were developed on glass slides and then incubated with Bacillus NOH and observed under light microscope, 24 h later (Figure 5). Light microscopic images revealing the biofilm reduction confirms the 24 h-dispersion kinetics.
Preliminary investigation of the anti-biofilm activity of B. subtilis strains
Bacterial probiotic defenses can act first and directly on pathogenic growth, targeting the adhesion and formation/ maturation of biofilms and indirectly by disturbing matrix cohesion via secretion of anti-biofilm substances. Because the B. subtilis species were the most efficient in biofilm biomass reduction, we further investigated their mode of action. Given the diversity of anti-biofilm defence modes, we first tested the influence of Bacillus on pathogen growth, using a growth inhibition test with B. subtilis CU1, B. subtilis var. natto, and B. subtilis NOH against A. naeslundii, S. mutans, S. sobrinus, and S. oralis separately (Figure 6). Among the three probiotics tested, only B. subtilis CU1 can inhibit growth of all four biofilm strains. Indeed, a clear inhibition zone formed around each pathogen strain upon exposure to this probiotic. In contrast, B. subtilis NOH and B. subtilis var. natto showed no inhibition of these four pathogens.
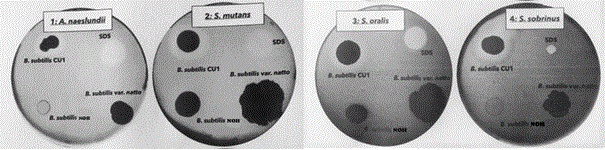
Figure 6: Growth inhibition test: antimicrobial activity of B. subtilis CU1, B. subtilis var. natto, and B. subtilis NOH strains using a modified agar well diffusion method against each oral pathogenic bacterium: A. naeslundii, S. mutans, S. sobrinus, and S. oralis.
Of note, A. naeslundii and S. sobrinus appeared to inhibit B. subtilis NOH growth, which had a limited or even absent growing zone with these strains compared to its growing zone on plates with S. mutans or S. oralis.
Prevention of biofilm formation
We speculated that an antibacterial effect as the only antibiofilm strategy would not be sufficient to destabilize an existing biofilm but could have a decisive impact on biofilm formation. To confirm the antibacterial effect of the B. subtilis CU1 strain compared to the other two Bacillus strains (NOH and natto), we tested the effects of different cell concentrations on interference with biofilm development (Figure 7).
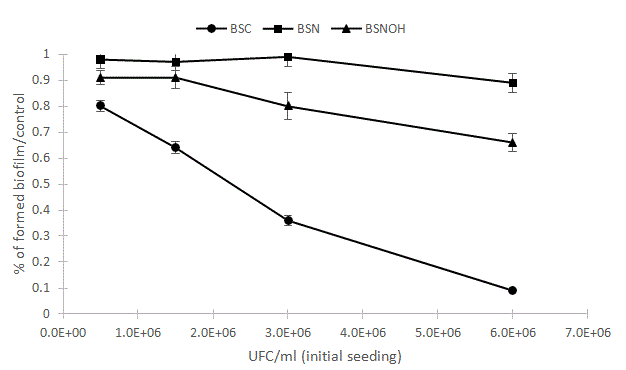
Figure 7: Effects of single probiotic strain on biofilm formation. The pathogenic strains S. mutans, S. sobrinus, S. oralis and A. naeslundii were mixed at a dose of 5 � 106 (CFU/ml) with or without Bacillus subtilis NOH (BSNOH), natto (BSN), and CU1 (BSC) probiotic strains at doses of 0.5, 1.5, 3 and 6.106 CFU/ml. After 24 hours of co-incubation at 37°C, the biofilm was stained with safranin 0.25% and quantified using spectrophotometry at a wavelength of 492 nm. Error bars represent the standard deviations of the means of four experimental replicates.
Inhibition of biofilm development relies on several antimicrobial mechanisms, including direct growth inhibition, reduction of microbial adhesion, and suppression of pathogen fixation. Thus, the dose effect of the Bacillus strain on biofilm synthesis appeared to confirm a direct growth inhibition of all biofilm strains by the CU1 strain, which completely prevents the construction of the biofilm at a high cell concentration (Figure 7). Likewise, these results support the implication of different mechanisms for the two other Bacillus strains, which were less effective in countering the construction of biofilms.
Pre-formed biofilm disruption ability of B. subtilis NOH and conditioning media
We sought to determine whether biofilm degradation by this strain was driven by the cells or extracellular compounds produced in the supernatant. Our results indicated that both the cell-free supernatant and cell culture exerted biofilm degradation activity at same levels of intensity after 24 hours of incubation (Figure 8). These findings suggest that this strain can secrete one or more anti-biofilm molecules.
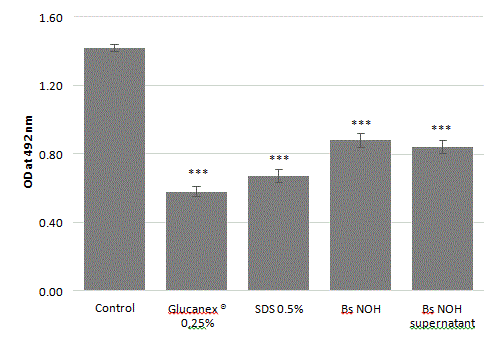
Figure 8: Amount of biofilm degradation by probiotic cells or cell-free supernatant against A. naeslundii, S. mutans, S. sobrinus and S. oralis mature biofilm. Reference positive controls are SDS (sodium dodecyl sulfate 0.5%) and Glucanex® (Lysing Enzymes from Trichoderma harzianum 0.25%). Error bars represent the standard deviations of the means of 5 independent experiments (*** indicates a significant difference from the control condition, p<0.001).
This Bacillus species is recognized for its production of lytic enzymes, which can serve as biofilm-degrading agents [49]. We did attempt supernatant purification by means of protein precipitation with ammonium sulfate followed by dialysis. Some of these purified fractions were then tested on a mature biofilm, but the results were not conclusive. Indeed, the anti-biofilm activity of the purified fractions was not reproducible and lacked stability, unlike Bacillus culture, which maintained a more constant activity.
We also sought to explore which B. subtilis NOH factors might contribute to biofilm degradation and tried some modifications to the culture media to stimulate or stabilize the production of anti-biofilm molecules by B. subtilis NOH. For instance, we evaluated solid-state fermentation on soy cake, which increases Bacillus protease production, but found no improvements compared to submerged fermentation [50].
Surfactants are an important class of chemical compounds with antimicrobial, anti-biofilm, and anti-adhesive properties [51]. Bacillus is one of most competent microbial genera in producing biosurfactants and a broad spectrum of lipopeptide biosurfactants [52]. For this reason, we also used the rapid “drop-collapse assay” to detect biosurfactant production by Bacillus subtilis liquid cultures (Figure 9).
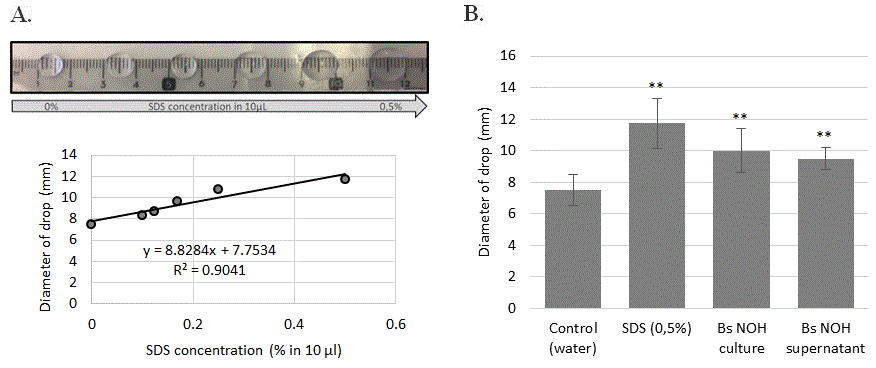
Figure 9: Tensioactive property of B. subtilis NOH supernatant. (A) SDS dose effect (%) on diameter drop. (B) Drop diameter of B. subtilis NOH supernatant and culture. Error bars represent the standard deviations of the means of 4 independent experiments. ** means statistical difference (p<0.01) between the marked sample and control.
The results presented in Figure 8 shows that with a control synthetic surfactant (sodium dodecyl sulfate), there is a direct correlation between droplet diameter and surfactant concentration. The comparison of the probiotic-exposed culture with this control indicated the presence of biosurfactant in whole and supernatant-derived B. subtilis NOH culture, corresponding to the effect of 0.275% SDS.
In the oral cavity, biofilm construction mediated by bacterial glucosyltransferases leads to complex 3D architectures in which insoluble α-1,3-linked glucans are crucial for structural integrity. The highly insoluble, branched, and structurally rigid glucans embed the cells, contributing to the scaffolding of the EPS matrix, and are responsible for a wide range of infectious diseases, including dental caries. The rigid extracellular matrix allows for bacterial adhesion-cohesion and drug tolerance, making biofilms difficult to treat using conventional or natural antimicrobial monotherapy [53].
Before biofilm maturation completes, however, the different stages of its formation are more or less controllable by different chemical or microbiological (probiotic) agents that can act on pathogen growth, adhesion, fixation, and signaling pathways (e.g. quorum sensing) [54-56]. The effective intervention of beneficial microorganisms against biofilm construction is well documented for many probiotic genera such as LactoBacillus, Streptococcus, Bacillus, and Saccharomyces (Table 1).
To confirm the potential of these probiotics, we sought to assess the probiotic disruption of the three-dimensional protection of an established biofilm to increase access to its internal structures for other treatments. To establish a representative in vitro model of a strong mixed-species biofilm, we used S. mutans together with A. naeslundii, S. sobrinus and S. oralis to intensify expression of both gtfB and gtfC [28]. This highly heterogeneous architecture may create niches that are essential for biofilm virulence and is adapted for the study of probiotic capacity for disrupting it.
Bacterial defenses can act first and directly on pathogenic growth, targeting the adhesion and maturation of biofilms and indirectly by disturbing matrix cohesion via secretion of antibiofilm substances. Indeed, some bacteria can produce several enzymes capable of disrupting the protection offered by the biofilm matrix by modifying its architecture. Our results show that among different genera of the probiotics tested, only those of the Bacillus genus exerted significant biofilm degradation abilities, reaching up to 40% for the best performing strain, which was isolated from a traditional Japanese fermented food (natto). This strain has been formally identified through gyrA sequencing and belongs to the B. subtilis species [37]. The identified degradation rate was significant and approaches the destructive powers of the positive enzymatic (Glucanex®) and surfactant (SDS) controls, which both reached ~55% biofilm degradation. Light microscopic images revealing the biofilm reduction confirms the 24 h-dispersion kinetics.
The anti-biofilm action of the NOH strain starts quickly and is almost complete after 5 hours of probiotic treatment. However, the effective dose, expressed as cells density, required to achieve notable matrix reduction, is around 108 CFU/ml.
Other strains, including L. rhamnosus GG, S. boulardii, S. salivarius K12, S. dentisani, and L. gasseri, had no significant impact on the mature biofilm. In addition, strains such as L. reuteri, L. salivarius, L. paracasei, and L. acidophilus, which have had a well-documented positive impact on biofilm “formation”, did not affect mature biofilm disorganization but rather promoted a slight consolidation [21,23-25,34-36]. These probiotic effects can be more subtle than pure destruction and could, for instance, influence matrix composition; indeed, in our in vitro model, we found that these probiotics could integrate into the biofilm.
These results are not surprising because the only publications describing microorganisms that can dismantle a mature oral biofilm most often mention the extracellular production of glucanohydrolases, such as mutanase (α-1,3-glucanase) and dextranase (α-1,6-glucanase), which can hydrolyze water-insoluble glucan [57]. In fact, we found that the partial biofilm matrix breakdown by B. subtilis NOH, isolated from natto, resulted from exocellular activity, as demonstrated by the activity of the acellular supernatant. Unlike the B. subtilis CU1 strain, the antibiofilm activity of B. subtilis NOH is not supported by antibacterial properties against pathogenic microbes.
However, biofilms change the game by providing microbes with greatly increased protection from antimicrobials. Therefore, biofilm dispersal agents, that deprive the pathogens its protection, which is rather the strategy of the Bacillus NOH strain, will be able to complement the anti-pathogenic activity of Bacillus CU1. Enzymes have been proven to be effective for the degradation of the EPS of the biofilms. The mechanism by which enzymes destroy the physical integrity of the EPS is through weakening the proteins, carbohydrate and lipid that make up the structure of the EPS through the degradation process [58].
Bacillus subtilis can produce different types of biofilm matrixdegrading enzymes, such as proteases, dextranase, levanase, DNAse and others [57,59-62]. However, the nature of the antibiofilm agents secreted by the Bacillus NOH strain could not be established through conventional fractional purification trials with ammonium sulfate, which we conducted to explore enzymatic activities. Otherwise, the genus Bacillus is known for its ability to produce biosurfactants, and similar molecules, such as lipopeptides, can also destabilize a biofilm by SDS-like detergent effects [51]. Likewise, bio-surfactant detection tests indicated the presence of tension-active molecules in B. subtilis NOH cultures.
In conclusion, knowing that to eradicate biofilms, it is recommended to act by targeting both functionally and structurally extracellular polymeric matrix, the combinatorial treatment strategy with Bacillus strains CU1 and NOH will be interesting. Indeed, dual-targeting treatment that is both degrading and antimicrobial, by the combination of B. subtilis NOH with an anti-bacterial probiotic partner such as B. subtilis CU1, could dismantle the matrix scaffold and expose pathogenic cells for efficient killing. This pairing would be expected to simultaneously cause cellular dispersion and the physical collapse of the entire bacterial structure. B. subtilis NOH represents a promising candidate for an oral care product, either alone or in such a combination, but more research is needed to confirm its probiotic qualities and determine the nature of the molecules it secretes.
We would like to thank the “Wallonie-Bruxelles federation” for the funding of this research.
Citation: Van Vlasselaer M, Dillemans M, Van Nedervelde L (2021) Disruption Potential of a Multi-species Oral Biofilm by Probiotics: Preliminary Investigation of a New Bacillus subtilis Strain. J Prob Health. 9:233.
Received: 01-Feb-2021 Accepted: 15-Feb-2021 Published: 22-Feb-2021 , DOI: 10.35248/2329-8901.21.9.233
Copyright: © 2021 Van Vlasselaer M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : We would like to thank the “Wallonie-Bruxelles federation” for the funding of this research.