Journal of Food: Microbiology, Safety & Hygiene
Open Access
ISSN: 2476-2059
ISSN: 2476-2059
Research Article - (2022)Volume 7, Issue 7
The diversity of microflora was determined to assess the effectiveness of decontamination treatments on pasteurized meat carcasses at a large meat packing plant in Canada. The main objectives were to characterize the bacterial diversity surviving in the heat-treated and DNase-I-treated samples over the untreated samples of pasteurized meat carcasses. Cultivation based-methods were combined with nested PCR-DGGE fingerprinting methods to quantify low numbers of bacterial survival in each sample. Using the DGGE reference marker, seven genera (Pseudomonas, Staphylococcus, Propionibacterium, Chryseobacterium, Flavobacterium, Ralstonia, Paenibacillus) were detected by both DGGE and cultivation-based method. Three species (Streptococcus salivarius, Micrococcus luteus and Leuconostoc mesenteroides) were exclusively found in pure cultures with cultivation-based method. Over fifteen genera were found by PCR- DGGE using both the DGGE marker strains and bands in real samples, indicating the highest diversity determined by this technique. By contrast, the highest quantity of species was detected by cultivation-based method (29 and 88 in heat 34 treated and untreated samples, respectively). Five E. coli isolates in the family of Enterobacteriaceae were detected in untreated samples with plating methods demonstrating the usefulness of processing meat samples with decontamination and heat treatments. However, most of the unidentifiable species or genus by cultivation- based were almost detected by PCR-DGGE, confirming the effectiveness of combining both culture-dependent and independent methods to completely profile bacteria in food samples.
Meat carcasses; Cultivation-based method; Nested PCR-DGGE; Decontamination treatment; Pasteurization method
The quality and safety of meat offered to retailers and consumers by standard meat plants remain a high concern in developed countries. Therefore, several decontamination treatments are usually applied on meat carcasses to eliminate the microbial contamination sources. For instance, in North American meat plants, various chemical or physical decontamination treatments had been used to reduce the numbers of spoiled and pathogenic bacteria on beef carcasses in concordance with North American regulatory requirements [1-4]. An investigation for a control over microbiological contamination in a large beef packing plant in Canada revealed the presence of few numbers of E. coli (<1 cfu/1000 cm2) mainly during the breaking process on cooled meat carcasses [5]. This study reported that spraying the unviscerated carcasses with 5% lactic acid reduced the numbers of aerobic microbes by about a log unit, but with subsequent carcass dressing operations, a second treatment with 5% lactic acid, pasteurizing and carcass cooling had no substantial effect on the number of aerobic microbes on cooled beef carcasses. Nowadays, meat microbiologists usually found it necessary to consider the number as well as the presence of bacteria on raw meat for a better control of its contamination sources. Quid the meat plants and retailers in developing countries such as Africa ones where the costs of laboratory equipment are beyond the reach of industrials? Quid the level of microbiological contamination sources of raw meat in such regions where food safety and quality are often misunderstood or ignored? Thus, determination of molecular markers that characterize the microbial trend and biohazard of a critical control point in the food manufacturing process would help to enhance local foods safety and quality of meat in developing countries. Techniques that might be accurately and routinely used in an industrial level in developing countries are conventional microbiological controls, but their main limitations and drawbacks are related to their time-consuming. Low costs and specific molecular techniques that have been validated may be an alternative for meat industries in developing African countries. Quantification of the bacterial diversity of food matrices samples (milk, cheeses, meats, etc.) had been successfully accomplished by combining culture-dependent and independent methods. Culture dependent methods consist of isolating and culturing microbes prior to their identification, while culture independent methods directly target the intrinsic molecules without cultivation of the microorganisms [6]. However, most of these are high costs and might be beyond the reach of African meat industries and research laboratories. One of the most promising and low cost could be the PCR-DGGE fingerprinting technique. It is a valuable tool to detect microbes in situ of food matrices samples [7]. It had been used in several times in meat samples to characterize the microflora mainly during storage. For this purpose, Brightwell et al., [10] characterized the microflora of peroxyacetic acid treated and vaccum-packaged beef stored for upto 18 weeks at -1.5°C by combining conventional microbiological methods and molecular fingerprints. PCR-DGGE and clone library methods were able to detect Carnobacterium sp. and Clostridium sp, in both samples, respectively, while the cultivation- based methods did not, but accurately determined the total counts of Enterobacteriaceae. Other studies characterized microbial populations in meats exposed in air and or preserved in vacuum- packaging stored under various chilled temperature regimes. In the same time, storage of fresh meats under chilled temperature has been investigated by studying spoilage bacteria during storage and their spoiling potential [8-14]. Furthermore, efforts of research have been agreeably noticed to identify the main bacteria responsible for spoilage in both meat processing plants and meat products [15]. Yang et al [5] previously quantified aerobes, coliforms and or E. coli on chilled carcasses that survive those treatments using both multiplex and real time PCR. Mainly, almost no E. coli were detected in pasteurized meat carcasses exceptthat failure during control of meat contamination, to the best of our knowledge, no studies have not yet related the identification of microflora that survives subsequent decontamination treatments in pasteurized meat carcasses. The identification of bacteria in that step may allow establishing a biomarker tracking any microbiological contamination from failure control. In this study, the cultivation-based identification and nested PCR-DGGE fingerprinting methods were combined to characterize the microflora of pasteurized meat carcasses that survive decontamination treatments by identifying the survival bacteria in heat-treated and untreated samples after pasteurizing treatment and also by evaluating the microbial diversity of such population in heat-treated, DNase-I-treated and untreated samples.
Meat sample preparation
Twenty Sponges moistened with 10 ml of 0.1 % peptone water were used to randomly swab 20 carcass samples in areas of 10,000 cm2 (mainly on the brisket, forequarters and hindquarters) at the federally inspected meat processing plant (Calgary, AB, Canada) and collect samples. Sponges were then kept in ice and processed immediately after arrival to the laboratory. Each wet sponge was stomached for 2 min and then, stomacher fluids were mixed in final volume of 200 ml of bacterial suspension. That mixture was divided in 8 sterile tubes of 25 ml which were centrifuged at 10,000 g, 4°C, for 10 min. Then, the pellets were combined and re- suspended together in 10 ml of 0.1% peptone water which was the working bacterial suspension and kept at -80°C for immediate uses.
Isolation of bacteria
For aerobes and anaerobes isolation, a 10-fold dilution was prepared from 0.5 ml of working suspension by mixing 100 μl of that solution with 900 μl of peptone water. Then, the mixture was used to incubate 10 Trypton Soya Agar (TSA, Oxoid, ON, Canada) plates. Two parts of 5 TSA plates were used to recover aerobes and anaerobes. Each plate was inoculated with 100 μl of diluted suspension. 5 labelled aerobe plates were incubated aerobically at 25°C for 48 h and the other 5 labelled anaerobe plates were kept in anaerobic incubator jars using Oxoid AnaeroGen sachet (Oxoid) at 25°C for 48 h. Enumeration of spore-forming bacteria was monitored by heating 1.5 ml of the previous working suspension at 80°C for 20 min. 0.5 ml of suspension was used to prepare a 10-fold- dilution. Then, 0.1 ml portions of that dilution were spreaded on 10 plates of TSA. 5 plates were then incubated aerobically at 25°C for 1-3 days and 5 other plates incubated anaerobically using Oxoid AnaeroGen sachet (Oxoid) at 25°C for at least 10 days to enable growth of spore-forming bacteria.
Isolates identification
Conventional microbiological tests were performed to presumptively identify 100 viable colonies isolated from each aerobe and anaerobe plates. For this purpose, they were randomly selected for Gram staining, cellular arrangement observations, oxidase, catalase, glucose utilization and motility tests. Isolated bacteria are grouped as follow:
• Group I included Gram negative rod-shaped and aerobe bacteria; oxidase negative.
• Group II included oxidase positive.
• Group III included Gram positive anaerobe (facultative or obligate) bacteria, oxidase and catalase negative.
• Group IV included Gram-positive aerobe bacteria, oxidase negative and catalase positive.
• Group V included those oxidase and catalase positive.
• Group VI included Gram-positive anaerobe obligate bacteria, oxidase negative and catalase positive.
Conventional biochemical tests were used to accurately identify the isolates in each group using Analytical Profile Index (API) test strips 20E, 20 NE, 50CHB/E, 50 CHL and API STAPH (bioMerieux Canada, St-Laurent, Quebec) following the manufacturer’s instructions. API 20E and API 20NE test strips were used to identify isolates in Group I and II, respectively, according to the manufacturer’s instructions, with incubation of the strips at 35 and 29°C, respectively. Bacterial suspensions in saline solution or in AUX medium plus NaCl at a 1% (wt/vol) final concentration were used as the inocula for API 20E and API 20NE kits, respectively. Examination of the strips was conducted after 24 and 48 h. API 50 CHL was used to identify isolates in Group III according to the manufacturer’s instructions after cultivation of bacteria in MRS agar (Oxoid, ON, Canada) at 30°C for 48 h. Examination of the strips was monitored after 24 and 48 h at 30°C. API 50 CH B/E and API Staph tests were monitored to identify isolates in Group IV and V according to the manufacturer’s instructions at 29 and 35°C except that examination of the strips was conducted after 1, 2, 3 and 7 days. Then, microscopic observations of Bacilli spore- forming were checked on phase-contrast microscopy (Olympus, Olympus America, Mississauga, ON, Canada) to confirm whether they belong to the family of Bacillaceae. Then, all API profiles were identified using API Database version 4.1 (Apiweb; bioMerieux) [15,16].
Isolation of specific bacteria by plating methods and observation by phase-contrast microscopy for heat-treated samples
Bifidus Selective Medium agar (BSM, Sigma, Missouri, USA) was used for identification of presumptive bifid species after adding the BSM supplement (Sigma, Missouri, USA) freshly dissolved in 5 ml sterile water. Plates were analysed after 7 days of incubation at 37°C and the genus examination was realized using microbiological tests (catalase, oxidase,Gram coloration, growth conditions). Identification of Brochotrix thermosphacta bacterium was monitored by incubation for 7 days on STAA agar base (Oxoid) plates at 25°C. Bacilli spore-forming were checked on phase-contrast microscopy (Olympus) by observing endospores. Then, cells were incubated on API 50 CHB to determine the species similarity as previously described.
DNA extraction
The remaining working suspension of bacteria from carcass product swabs (8 ml) and the heated sample (1 ml) were used to perform DNA extraction. Two portions of 4 ml of the working suspension were separately used for bacterial DNA extraction. One portion was treated with 10 U DNase I (1 mg for 10 U, Sigma) prior to DNA extraction and the second portion was kept untreated. After treatment with DNase I, the first portion was heated at 65°C for 10 min to inactivate the DNase [17,18]. Then, DNA extraction was performed for untreated portion, portion treated with DNase I as well as the heated sample using the Fast DNA Spin kit for Soil (MP Biomedicals, Fountain Parkway, USA) according to the manufacturer’s instructions. The purity of DNA of each sample was determined using the ratio of A260/A280 by spectrophotometry (NanoDrop ND-1000, Nanodrop Technologies, Rockland, DE, USA).
Design of PCR-DGGE reference marker strains
Eighteen colonies were randomly selected from each group of isolates (groups I to VI and heat-treated sample) to extract DNA for DGGE analysis and comparison with cultivation-based methods. Twelve of them were selected and pooled as DGGE reference marker strains. The bands corresponding to each reference marker strain on the gel is identified to known taxon. DNA fragments from the samples having a same location as a reference marker strain in the DGGE gel is assigned to the same genus.
PCR-DGGE fingerprinting analysis
Total DNA (10 ng/μl) of each of all samples including reference marker strain was used as a template to generate the amplicon for PCR-DGGE analysis using nested PCR. The nested PCR was performed by amplifying the 1.5 kb product targeting the full length 16S Rrna gene with an universal bacterial primer pair 27F (5'- AGAGTTTGATCMTGGCTCAG-176 3') and 1492R (5’-TACGGYTACCTTGTTACGACTT) as previously described by Chen et al. [19]. The nested PCR conditions were: an initial denaturation for 5 min at 94°C; 30 cycles of 94°C for 30s, 58°C for 30 s, and 72°C for 90 s; and a final elongation for 7 min at 72°C. This PCR product was then diluted 10 times as a template to amplify a ~200 bp DNA fragment using HDA1-GC and HDA-2 primers [20]. These PCR conditions were an initial denaturation for 5 min at 94°C; 30 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 30 s; and a final elongation for 7 min at 68°C. After amplification, the products were subjected to DGGE analysis. All amplicons from 18 bacterial colonies were mixed and was loaded in one lane, while the individual amplicon in each real sample was loaded in the separate lanes of the same DGGE gel in order to determine the migration position of each band in the reference marker. DGGE was run on 1×TAE buffer (40 mM Tris-base, 20 mM acetic acid, 1 mM EDTA) with a 6% polyacrylamide gel with a 30-55% gradient gel using the Bio-Rad DCode universal mutation detection system (Hercules, CA) at 130 V, for 4 h. Then the gel was stained with 0.1% (vol/vol) ethidium bromide for 20 min. After washing, it was scanned using the FluorChem SP imaging system (Alpha Innotech, San Leandro, CA).
Cloning and sequencing analysis of DGGE bands
The sixteen separated bands generated from the real samples as well as the 18 reference marker strains were excised aseptically from the gel and transferred to diffusion 0.5 M ammonium acetate buffer, pH 8.0, containing ,10 mM magnesium acetate, 1 mM EDTA and 0.1% SDS (w/v). DNA fragments were extracted using the Qiaex II gel extraction kit (Qiagen Sciences, MD) according to the manufacturer’s instructions. Furthermore, the extracted products were reamplified using the same HDA1/HDA2 primer pair mentioned above. The fresh PCR products were then cloned into the TOP10 vector (Topo TA cloning kit; Invitrogen, Carlsbad, CA) using chemical transformation. Colonies were cultured on S-Gal medium (Sigma, St. Louis, MO) and randomly picked, and, from five replicates with insertions, plasmid DNA was extracted using a Plasmid Miniprep 96 Kit (Millipore, Billerica, MA). The sequence reaction was performed in a 10 μl total volume containing 0.5 μl of BigDye, 3.2 pmol of M13 primer (forward or reverse), 2.0 ul of 5 × sequencing buffer, and 2.0 μl of plasmid DNA as the template with the ABI 3730 sequencing system using the ABI Prism Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). All sequences were subjected to Blastn to determine the closest known taxon. The sequence composition of each band was compared using the RDP Classifier tool.
Population counts in meat carcass samples
Population counts (colony forming unit) are displayed in Table 1a. Aerobes obtained from untreated samples were the major populations with 3-log compared to anaerobes with about 2-log. In contrast, within heat-treated samples, mandatory anaerobes were the most dominant population (3.9 x 102 CFU/ml). A few amounts of aerobe bacteria were found from heat-treated sample.
| Type of samples | Isolates | CFU/ml |
|---|---|---|
| UT | Aerobes | 2.17 x 103 |
| Anaerobes obligate | 2.2 x 102 | |
| Anaerobes facultative | 3.1 x 102 | |
| Population (N) | Total counts | 2.4 x 103 |
| HT | Anaerobes obligate | 3.9 x 102 |
| Population (N) | Anaerobes facultative Total counts | a1.0 x 102 |
| 4.0 x 102 |
aEstimated number
Table 1a: Population counts (N) of isolates present in Untreated (UT) and Heat- Treated (HT) meat carcass samples, respectively. For UT sample, aerobes and anaerobes were recovered by incubating TSA plates in air and simultaneously in air (facultative) as well as in anaerobic jars free of oxygen (obligate) using Oxoid AnaeroGen sachet (Oxoid, USA) at 25°C. For HT sample, aerobes and anaerobes were recovered by incubating TSA plates in air and simultaneously in air as well as in anaerobic jars free of oxygen at 25°C, respectively.
Identification of isolates using culture-based methods
The isolates from untreated and heat-treated samples were identified using conventional microbiological and biochemical methods: Gram staining, oxidase and catalase tests, glucose utilisation, microscopic observations and phenotypic numerical classification test such as API strips (Tables 1a and Table 2). Six (6) groups of isolates from the untreated sample were reported according to the Gram staining characteristic, oxidase and catalase tests. The groups I and II were the most representative with 22 and 27% of isolates, respectively. 53 isolates (50 % in the total of population) are Gram-negative bacteria present in Groups I (24 isolates) and II (29 isolates). 54 isolates (50 % in the total of population) are Gram-positive bacteria present in the other groups. The untreated sample was mostly populated by aerobes with 86 isolates in a total population of 107 isolates (Table 1b).
| Group | No. of isolates | Frequency (%) | Gram negative | Gram positive | Aerobes | Anaerobes |
|---|---|---|---|---|---|---|
| I | 24 | 22 | 24 | - | 23 | 1 |
| II | 29 | 27 | 29 | - | 29 | - |
| III | 13 | 12 | - | 13 | 7 | 6 |
| IV | 13 | 12 | - | 13 | 13 | - |
| V | 11 | 10 | - | 11 | 11 | - |
| VI | 17 | 16 | - | 17 | 3 | 14 |
| Total | 107 | 100 | 53 | 54 | 86 | 21 |
Table 1b: Microbiological values obtained from isolates of Untreated (UT) samples.
| Group | Family | Number of isolates | |
|---|---|---|---|
| Untreated samples (UT) | Heat treated samples (HT) | ||
| IV, V, HT | Bacillaceae | 21 | 1 |
| I | Enterobacteriaceae | 18 | - |
| III | Lactobacillaceae | 15 | - |
| II | Pseudomonadaceae | 14 | - |
| VI, HT | Propionibacteriaceae | 6 | 27 |
| II | Sphingomonadaceae | 4 | - |
| IV, HT | Micrococcaceae | 2 | 1 |
| II | Flavobacteriaceae | 3 | - |
| II | Caulobacteriaceae | 1 | - |
| I | Neisseriaceae | 1 | - |
| II | Ralstoniaceae | 1 | - |
| III | Streptococcaceae | 1 | - |
| II | Vibrionaceae | 1 | - |
| VI | Listeriaceae | 1 | - |
| In all | Not identified | 18 | 20 |
| Total | 14 | 107 | 49 |
Table 2: Summary of the closest relative species identified by phenotypic numerical classification (API strip tests) in meat samples and incubation on BSM agar (Sigma, Missouri, USA).
Identification of isolates using phenotypic numerical classification method (API strips)
Each group of isolates was identified according to a suitable API strip characteristic: API 20E for Enterobacteriacea identification (Group I), API 20 NE for non-Enterobacteriacea (Group II), API 50 CH/L for Lactobacillus identification (Group III), API STAPH for Staphylococcus identification (Group IV, V and VI) and API 50CH/B for Bacillus identification (IV, V and VI). The closest relative species obtained for each group of isolates were reported in Table 2. From the untreated sample, groups I, II and III were dominated by Enterobacteriaceae (18 isolates), Pseudomonadaceae (14 isolates) and Lactobacillaceae (15 isolates), respectively. The population of Bacillaceae (21 isolates) was representative too in group IV and V. The group VI was mainly dominated by presumptive Propionibacteria (6 isolates) detected on bifidus selective medium agar (BSM, Sigma, Missouri, USA). The greatest microbial diversity was found in group II with 6 different families: Pseudomonadaceae (14 isolates), Sphingomonadaceae (4 isolates), Flavobacteriaceae (3 isolates), Caulobacteriaceae (1 isolate), Ralstoniaceae (1 isolate), Vibrionaceae (1 isolate). The main dominant species in this sample were: Pantoea sp (12 isolates) and Escherichia coli (5 isolates) in Enterobacteriaceae family, Pseudomonas fluorescens (11 isolates) in Pseudomonadaceae, Lactobacillus sp (13 isolates) in Lactobacillaceae, Aneurinibacillus aneurilyticus (16 isolates) and Bacillus sp (4 isolates) in Bacillaceae. Photobacterium damslae strain in Vibrionaceae and Brochotrix thermosfacta in Listeriaceae were particularly detected in groups II and VI, respectively (Table 2). In heat-treated samples, 27 anaerobes obligate isolates were presumptively identified as Propionibacterium sp on bifidus selective medium agar (Sigma). They were characterized as Gram-positive, nonsporing, nonmotile, pleomorphic rods. Although some strains may be relatively aerotolerant, they are basically anaerobes that produce propionic acid, acetic acid, and CO2 as their main fermentation products. Their optimal growth temperature is between 30 and 37ºC [21]. Sporing Bacillus sp and Staphylococcus spstrains were detected in that sample too.
Comparison of PCR-DGGE analyses of pure colonies with those of cultivation-based by designing DGGE reference maker strains
Based on the positions of each band in the reference lane from 18 DGGE marker strains (lane V), 12 strong bands (lane IV) were selected and pooled as a DGGE reference marker strain (Figure 1 and Table 3). Each band of the reference maker strain was sequenced and identified (Table 3) by determining the closest relative species obtained by Blastn Search on GenBank database at NCBI. By comparing the phenotypic identification methods of pure colonies (API strips and cultivation on media) with 18 DGGE marker strains, the identification of 3 colonies by cultivation-based was consistent to the DGGE marker at the genus level (Propionibacterium, Pseudomonas and Staphylococcus). However, 12 colonies were not identifiable by cultivation method while 3 colonies were unable by DGGE. Using DGGE reference marker, 7 genera (Pseudomonas, Staphylococcus, Propionibacterium,Chryseobacterium, Flavobacterium, Ralstonia, Paenibacillus) were detected by both DGGE and cultivation-based methods. 3 species (Streptococcus salivarius, Micrococcus luteus and Leuconostoc mesenteroides) were exclusively found in pure cultures by cultivation-based method (Tables 4 and 5) (Figure 1).
| Band # | ID (No and group) | DGGE and sequencing identification | API and plates identification | ||
|---|---|---|---|---|---|
| Closest relative species | Similarity | Closest relative species | Similarity | ||
| a | 22-I | Staphylococcus capitis | 98 | 1 NI | |
| b | 55-V | Chryseobacterium joostei | 99 | NI | |
| c | 5-VI | Streptococcus infantis | 99 | NI | |
| d | 49-II | Flavobacterium sp | 99 | Mannheimia haemolytica / | 86 |
| Pasteurella trehalosi | |||||
| e | 17-VI | Streptococcus sanguinis | 98 | NI | |
| f | A21 | Staphylococcus pasteuri | 98 | NI | |
| g | 23-III | Streptococcus salivarius | 99 | NI | |
| h | 7-IV | Ralstonia solanacearum | 96 | NI | |
| i | 55-III | Paenibacillus borealis | 97 | Lactobacillus crispatus | 100 |
| j | 57-I | Micrococcus luteus | 98 | NI | |
| k | ANO-HT 28 | Propionibacterium acnes | 99 | 2Propionibacterium sp | |
| (Presumptive) | |||||
| l | 45-III | Luteococcus sp. | 96 | Lactobacillus delbruecki ssp | 99 |
| Delbruecki | |||||
| 3? | 28-II | Pseudomonas trivialis | 98 | Pseudomonas fluorescens | |
| 3? | 36-IV | NI | NI | ||
| 3? | 39-IV | NI | NI | ||
| 3? | 8-V | Pseudomonas marginalis | 99 | NI | |
| 3? | ANF-HT 02 | NI | NI | ||
| 3? | ANF-HT 33 | Staphylococcus saprophyticus | 99 | Staphylococcus sp | |
1NI: Not identified with Blastn search, 2 Presumptive Propionibacteria obtained by cultivation on BSM agar (Sigma) and checked on phase-contrast microscopy for cells morphology; ANO-HT: Anaerobe obligate-heat treated sample; ANF-HT: Anaerobe facultative-heat treated sample. 3? Six bands were not used as reference marker strain.
Table 3: DGGE reference markers performed for bacterial identification and comparison with identified species by cultivation-based method.
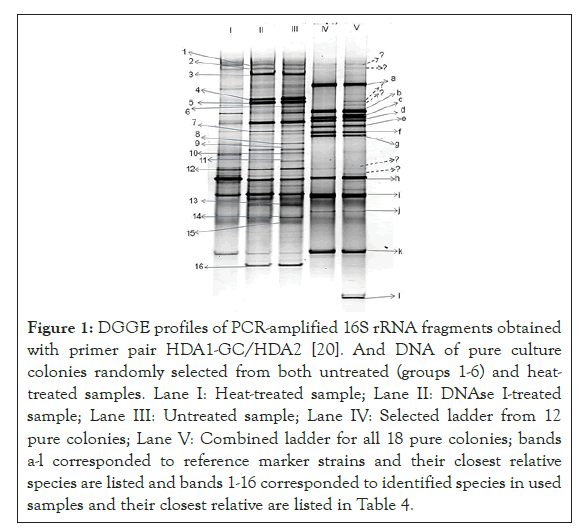
Figure 1: DGGE profiles of PCR-amplified 16S rRNA fragments obtained with primer pair HDA1-GC/HDA2 [20]. And DNA of pure culture colonies randomly selected from both untreated (groups 1-6) and heat- treated samples. Lane I: Heat-treated sample; Lane II: DNAse I-treated sample; Lane III: Untreated sample; Lane IV: Selected ladder from 12 pure colonies; Lane V: Combined ladder for all 18 pure colonies; bands a-l corresponded to reference marker strains and their closest relative species are listed and bands 1-16 corresponded to identified species in used samples and their closest relative are listed in Table 4.
| Band # | Description | Similarity | NCBI accession number |
|---|---|---|---|
| 1 | Pseudomonas aeruginosa strain AS1 | 97% | GU447237.1 |
| 2 | Halomonas sp. wp29 | 97% | AJ551106.1 |
| 3 | Pseudomonas fluorescens strain MR-6 | 100% | GQ906772.1 |
| 4 | Uncultured Verrucomicrobia bacterium | 96% | DQ446121.1 |
| 5 | Uncultured Bradyrhizobium sp. | 98% | HM996739.1 |
| 6 | Uncultured Actinobacterium | 100% | FN668331.2 |
| 7 | Proteobacterium BHI60-9 | 100% | AJ431217.1 |
| 8 | Uncultured Sphingobacterium sp. | 100% | FN668102.2 |
| 9 | Rhodococcus opacus gene | 96% | AB060974.1 |
| 10 | NIa | ||
| 11 | Uncultured bacterium | 97% | JF495433.1 |
| 12 | Mycoplasma sp. | 100% | FN421445.1 |
| 13 | Leuconostoc mesenteroides strain MBF2-1 | 98% | GQ456941.1 |
| 14 | Methanospirillum sp. | 100% | EU498392.1 |
| 15 | Butyrivibrio fibrisolvens | 98% | AM039822.1 |
| 16 | Algoriphagus sp. BW86-72 | 100% | FN395254.1 |
a NI: Not identified with Blastn search, although band #10 was present in heat-treated and untreated samples
Table 4: Sequence identification of the PCR-DGGE bands.
PCR-DGGE fingerprints of microflora in untreated, DNaseI-treated and heat-treated samples
The diversity and dynamic of microflora in heat-treated and DNaseI-treated samples were compared to those of untreated sample using PCR-DGGE fingerprint. To characterize the taxonomy of 16 bands selected from real samples, all of them were purified, cloned, sequenced and identified (Table 4). The following criteria were used to determine the taxonomy of each band: a 96% or higher match between the clone sequence and the GenBank data was considered to 260 represent identity at the species level, and a 90% to 95% match represented identity at the genus level, as given by the RDP Classifier online tool. From the sequences obtained from the 16 PCR-DGGE bands, 9 of them corresponded to the known species: Pseudomonas aeruginosa, Halomonas sp., Pseudomonas fluorescens, Rhodococcus opacus, Mycoplasma sp.,Leuconostoc mesenteroides, Methanospirillum sp., Butyrivibrio fibrisolvens and Algoriphagus sp. One sequence was identified only at the genus level (band 7 as Proteobacterium). Five sequences matched uncultured clones presented the following genera: Verrucomicrobia, Bradyrhizobium, Actinobacterium, Sphingobacterium and uncultured Bacterium. Compared bacterial profiles generated from three different treated samples, bands 3, 4, 5, 9, 11, 14 and 16 (Pseudomonas fluorescens, uncultured Verrucomicrobia bacterium, uncultured Bradyrhizobium sp. Rhodococcus opacus, uncultured bacterium, Methanospirillum sp. and Algoriphagus sp.) were absent in heat treated sample. Band 8 (Uncultured Sphingobacterium sp.) and band 10 (uncultured bacterium) were absent in DNAseI-treated sample. Band 6 (uncultured Actinobacterium) was only present in the sample without treatment (Tables 4 and 5). Based on the identification of species in each sample using the DGGE reference marker, Streptococcus salivarius (band G), Micrococcus luteus (band J), and Luteococcus sp (band I) were not detected at all (Table 5). By contrast, Staphylococcus capitis (band A), Chryseobacterium joostei (band B), Flavobacterium (band D), Raltsonia solanacearum (band H), Paenibacillus borealis (band I), Propionibacterium acnes (band K) were detected in each sample. Streptococcus infantis (band C) and Streptococcus sanguinis (band E) were only present in untreated sample while Staphylococcus pasteuri occurred in both DNAseI-treated and untreated samples. In a total of 74 known species found by DGGE in all samples, 18, 25 and 31 species were detected in heated, DNAseI-treated and untreated samples, respectively, while 29 and 89 species were identified in heat-treated and untreated samples, respectively, by cultivation-based method (Tables 3).
| Band # | Heated | DNAse I-treated | Untreated |
|---|---|---|---|
| 1 | + | + | + |
| 2 | + | + | + |
| 3 | - | + | + |
| A | + | + | + |
| 4 | - | + | + |
| 5 | - | + | + |
| 6 | - | - | + |
| B | + | ± | + |
| C | ±± | - | + |
| D | ± | + | + |
| E | - | - | + |
| 7 | + | + | + |
| F | - | + | + |
| G | - | - | - |
| 8 | + | - | + |
| 9 | - | + | + |
| 10 | + | - | + |
| 11 | - | + | + |
| 12 | + | + | + |
| H | + | + | + |
| I | + | + | + |
| 13 | - | + | + |
| J | - | - | - |
| 14 | + | + | + |
| 15 | + | + | + |
| k | + | + | + |
| 16 | - | + | + |
| l | - | - | - |
| *? (12) | + | + | + |
| ? | ± | ± | ± |
| ? (6) | - | - | + |
| ? (5) | - | + | + |
| ? (1) | + | + | + |
| ? (2) | + | + | + |
| Total of species present in | 18 | 25 | 31 |
* Six bands not selected as reference marker strain; faint bands (±) were considered as present; very faint bands (±) were considered as absent.
Table 5: PCR-DGGE bands of both tested samples and reference marker strains. Presence (+), faint (±), very faint (±±) and/or absence (-). Lane I: Heat treated sample; Lane II: DNAseI-treated sample; Lane III: Untreated sample; Lane IV: Selected ladder from 12 pure colonies; Lane V: Combined ladder for all 18 pure colonies.
This novel study on the microbiological quality control data collected after decontamination treatments on pasteurized meat carcasses was performed to provide more information about the diversity of the microflora. Thus, cultivation methods were able to recover both aerobes and anaerobes in untreated samples. Groups (six) of isolates were determined in that sample based on their Gram-stain, catalase, oxidase and glucose utilization characteristics. Bacillaceae, Enterobacteriaceae, Lactobacillaceae, Pseudomonadaceae, Propionibacteriaceae, Sphingomonadaceae, Micrococcaceae, Flavobacteriaceae, Neisseriaceae, Pasteurellaceae, Ralstoniaceae, Streptococcaceae, Vibrionaceae and Listeriaceae were prominent in that sample. Bacillus sp, Pantoea sp, Escherichia coli, Lactobacillus sp, Propionibacterium sp, Sphingomonas sp, Staphylococcus sp and Streptococcus sp were the most dominant species identified by cultivation method. One of the major findings in this sample was the occurrence of E. coli identified for 5 isolates in the Enterobacteriaceae group. The presence of E. coli in the sample could be justified by the fact that chemical decontaminants sprayed on the carcass sides could not reach zones where E. coli continue growing [22]. These findings agreed with a previous study which reported that most cooled carcasses carry E. coli at numbers <1 cfu/10,000 cm2, but that product can be contaminated with small numbers (<1/100 cm2) of E. coli during carcass breaking. Bacillus sp strains are widespread in meat and meat products and their occurrence in our samples (untreated and heat-treated) is not surprising. Many of them showed spore-forming ability which promotes the survival of members of this genus during food processing treatments such as milk, juice and meat pasteurization [23,24]. In the other hand, Pseudomonas sp was the one of the main genera found in untreated samples. Particularly, Pseudomonas sp are known to be involved in the spoilage of meat stored at chilled temperature as they are regarded as psychrotrophic bacteria. The occurrence of Sphingomonas paucimobilis (4 isolates) in this group could be explained by the presence of some biological pollutants in the meat plant air. This species is known to play an important role in bioremediation (the use of biological agents to remove pollutants from the environment) as they break down the polycyclic aromatic hydrocarbons [25]. Some fastidious and specific bacteria were detected by cultivation on specific media and observed on phase-contrast microscopy (Olympus). Thus, one (1) isolate was identified as Brochotrix thermosphacta in the untreated sample by cultivation on STAA medium (Oxoid). This strain is a spoil bacterium usually found in many meat samples and its occurrence is regularly increased during storage mainly on packaging conditions [26]. Furthermore, the growth of 33 isolates in Bifidus Selective Medium agar (Sigma) for both untreated and heat-treated samples is interesting findings as Propionibacteria are difficult to find in meat sample for quality control by cultivation methods. The characteristics of this group of bacteria are consistent to these isolates: they are Gram-positive bacteria, catalase positive and oxidase negative, pleomorphic rods with characteristic V or Y configurations, naturally present in rumen cows, widespread in milk and are indicators for skin contamination of human origin and can resist to heat at 90°C for 10 min [27,28]. These bacteria were presumptively identified from 6 and 27 isolates in both untreated and heat-treated samples, respectively. Then, cultivation- based methods are not enough to confirm their occurrence. Culture-independent methods are now emerged to accurately recover microbes within food samples without any cultivation. PCR-DGGE fingerprinting is one of the promising biomolecular method for detecting bacteria and studying microbial population dynamics in food samples. It relies on electrophoresis separation of PCR products of different DNA fragments of similar length. However, one of the limits of PCR-DGGE is due to the fact that it is not a quantitative method because the band intensity is not well correlated with plate counts [29,30]. Nevertheless, this technique could contribute to complementary information to plate counts method. Prior to DGGE analysis, nested PCR was used to increase the sensitivity as a low abundance of microorganisms was expected in some samples such as ours. PCR-based DGGE technique had been extensively and successfully used to monitor the change in microbial composition during different storage conditions. So, it may be a helpful tool to determine the fingerprints of few microbial populations surviving the decontamination treatments of chilled meat samples before storage. A DGGE reference marker strain was first designed using selected DGGE bands from the profiles obtained by amplifying the DNA of randomly selected pure cultures of isolates from heat-treated, DNAseI-treated and untreated samples. This bacterial marker could be a quick method to monitor the presence of known microorganisms (viable, uncultured, viable but not cultivable and dead microorganisms) occurring in each sample. Heat treatment used as a pasteurization process in many food samples carried less microbial diversity and, then, remain the method of choice to eliminate the bacterial contaminants and pathogens in food. In the present contribution, heat treatment was applied to sample as a pasteurizing process to remove psychotropic and mesophilic spoilage bacteria remained in the chilled meat sample. Among 18 bacteria found in heat-treated samples, only 2 bacteria (bands 8 and 10) were uncultured and the viable bacteria were fewer than in DNAseI-treated sample. According to this result, DNase I treatment was effective enough to remove dead cells as shown in band 10 (Lane II). In heated-treated and untreated samples, band 10 (Lane I and III, respectively) showed uncultured bacteria which were not identified with blastn search, confirming the role of DNase I treatment to remove dead cells. Heating process is a physical treatment during which dead cells are expected to undergo cell lysis or at least lost integrity of cellular boundaries. By contrast, the DNAse-I treatment is an enzymatical treatment that was applied to samples to hydrolyze DNA from dead cells and subsequently detect the presence of living bacteria by conventional PCR. This method had been proven to successfully detect specific pathogens or possible live microbial contaminants in food samples [31]. Of the 25 bacteria found in DNAseI-treated sample, only 2 bacteria (band 4 and 5) were uncultured compared to untreated sample suggesting the effectiveness of DNAseI treatment to detect only live bacteria. DNAseI might not have a damaging effect in DNA of live/intact cells, its treatment to discriminate viable and non-viable cells was based on the assumption that when the cell is dead, it looses its membrane property of selective permeability and will facilitate the passive movement of DNAseI into the cells [32]. Similarly, a biological based-PMA (Propidium monoazide) treatment to discriminate between live and dead cells is being increasingly used by food microbiologists to detect pathogen or poisoning bacteria in foods as its combination with real time PCR was reliably demonstrated [33]. Furthermore, more than 15 genera were found by PCR-DGGE using both the DGGE marker strains and bands in real samples, indicating the highest diversity determined by this technique. By contrast, the highest quantity of species was detected by cultivation-based method (29 and 88 in heat-treated and untreated samples, respectively). However, most of unidentifiable species or genus by cultivation-based were almost detected by PCR-DGGE, confirming the effectiveness of combining culture-dependent and independent methods to completely profile bacteria in food samples [34].
Decontamination treatments had been proven to effectively eliminate microbial contaminants and spoilage bacteria in meat carcass samples. Although the PCR-DGGE method demonstrated its effectiveness on microbial diversity assessment, this study confirmed particularly the presence of few numbers of E. coli in decontaminated raw meat sample, exclusively detected by cultivation-based method. Therefore, a rapid diagnostic and quantification method is needed to improve selective detection of generic and verotoxigenic E. coli (VTEC) isolates in raw meat samples. For these reasons, current programmes are being investigated for detection of such microorganisms in meat products Therefore, important efforts of analysis and research on raw meat quality should be conducted for public health safety in Africa.
We address grateful thanks to the Albertan Livestock and Meat Agency (ALMA), Canada, for its financial support. BN, a former Visiting Scientist Fellow under the terms of the Visiting Fellowships in Canadian Government Laboratories Program, was awarded by the Natural Sciences and Engineering Research Council of Canada (NSERC) to undertake this project. Authors reserve a deep recognition to RABIOTECH (West African Biotechnology Network) headquartered at Ouagadougou (Burkina Faso) to financially support the publication costs. They finally take this opportunity to pay a posthumous tribute to Dr. Colin Gill (RIP) for his wide contribution as a world-class senior food microbiologist.
This study was funded by Alberta Livestock and Meat Agency (ALMA) and Natural Sciences and Engineering Research Council of Canada (NSERC) under the respective contracts No: 2011R025R and 402034.
BN carried out all experiments at Lacombe Research Centre (Lacombe, AB) and drafted the manuscript, XY and LLG coordinated the molecular technics experiments, KAS is a veterinarian who approved the methodology, MHD and IN critically reviewed the manuscript, AT and ATG are emeritus Professors who completed the design of the manuscript, COG (RIP) conceived, designed and was awarded the project’s funds.
Citation: Ndoye B, Yang X, Guan LL, Sylla KA, Dicko MH, Ndoye I, et al. (2021) Diversity of Microflora Surviving Decontamination Treatments of Pasteurized Meat Carcasses Assessed Using Cultivation-Based and Nested PCR-DGGE Fingerprinting Methods Poblational Study. Food Microbial Saf Hyg. 6:149.
Received: 17-Jun-2021 Accepted: 01-Jul-2021 Published: 08-Jul-2021 , DOI: 10.35248/2476-2059.21.6.149
Copyright: © Ndoye B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : This study was funded by Alberta Livestock and Meat Agency (ALMA) and Natural Sciences and Engineering Research Council of Canada (NSERC) under the respective contracts No: 2011R025R and 402034