Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Research Article - (2016) Volume 4, Issue 2
N-methylpyrrole (Py)-N-methylimidazole (Im) polyamide (PI polyamide) are increasingly used in basic and applied biomedical research. Although β-alanine (β) substitutions in 8-ring PI polyamides have been well analyzed, efficacy of double β substitutions for lengthened DNA minor groove recognition has not been elucidated in vivo. Here, we show effectiveness of double β substitutions in PI polyamides to retain high affinity of specific lengthened DNA binding and suppress target gene expression in cells. Initially we synthesized four 12-ring PI polyamides targeting the AP-1 site within MMP-9 gene promoter, 1-4, including a β/β pair (3) and two adjacent Py/β and β/Im pairs (4) and investigated the binding kinetics with oligoDNA containing preferred sequence (5’-AGTCAGCA-3’) by surface plasmon resonance assays and MMP-9 mRNA expression in MDA-MB-231 cells. The PI polyamides 3 and 4 showed high affinities of target DNA binding (KD=4.33×10-8) and (KD=5.00×10-8), respectively and significant down regulation of MMP-9 expression. We then considered adjacent Im/β and β/Im pairs should solve the problem of repeating GC sequence. A PI polyamide 5 targeting the E2F site, 5’-TTGGCGC-3, within the MYCN promoter was synthesized and tested binding affinity and MYCN suppression in MYCN amplified CHP134 human neuroblastoma cells. The PI polyamide 5 showed high affinity (KD=3.07×10-8) binding of the target sequence and significantly suppressed MYCN mRNA expression. Those results demonstrated a possible use of the adjacent double β substitutions for 12-ring PI polyamides, particularly in G/C rich regions and suggested substitutions of β springs in PI polyamides may extend applications for in vivo biomedical research targeting lengthened genomic DNA.
Keywords: PI polyamide, Minor grove binder, β-alanine, Sequence recognition, MMP-9, MYCN
Ever since the discovery of distamycin, a natural product antibiotic, and it’s surprising binding interaction with DNA to bind minor groove of DNA in a nucleotide-specific manner [1], tremendous efforts have been made to develop synthetic mimics with similar chemical properties for applications requiring specific DNA recognition in the past decades. Hairpin N-methylpyrrole (Py)-N-methylimidazole (Im) polyamides (“PI polyamide”) are a class of such molecules that bind the minor groove of double-stranded DNA and demonstrate some of the most promising results to-date. PI polyamides often appear in a configuration in which two oligoheterocyclic units are ligated by an aliphatic linker [2], and the intramolecular stacking of those face-toface heterocycles lead to a compact hairpin conformation Heterocycles in PI polyamides can chemically distinguish Watson-Crick base pairs via intermolecular hydrogen bonding between the amide groups in the polyamide molecule and nucleobases in the minor groove of DNA, with Im/Py recognizing G/C and Py/Py to A(T)/T(A) pairings [3] Their ability to differentiate nucleobases in a sequence-specific manner enable PI polyamides to interact with DNA in a variety of biological applications, such as the modification of transcription regulation [4-8]. However, there is a limit for the molecular structure as the lengthening of heterocycle chains in PI polyamides increase its structural rigidity and consequently alter the complementary curvature required for minor groove binding. The over-curved structure, as a direct result of chain lengthening, can lead to the detachment of a PI polyamide from its target DNA sequence [9]. As such, flexible subunits in Im-Py chains, for instance aliphatic β-alanines (β), are commonly introduced to alleviate the effect of chain over curvature [9]. In this context, the elongation of motif recognition up to eight base pairs led to the design and synthesis of a series of flexible PI polyamides including a β/β pairing, and was met with success [10]. Our previous report of a β/β-containing PI polyamide 3 (Figure 1) targeting the 5'-AGTCAGC-3' sequence in the matrix metalloproteinase 9 (MMP-9) promoter adjacent to the activator protein 1 (AP-1) binding site showed remarkable inhibition of MMP-9 expression in MDA-MB-231 human breast cancer cells [10]. However, a β/β pair recognizing only A(T)/T(A) base pairs also limited the range of target sequences for PI polyamides. Dervan et al. previously showed that β/Py and β/Im pairs, recognizing A(T)/T(A) and G/C respectively, could achieve specific 6-bp recognition of 8-ring PI polyamide in vitro [11]. The incorporation of two β moieties into a polyamide in the form of not only β/β pairs, but two discrete X/β (X=Im or Py) pairs, are beneficial for expanding target recognition while retaining the significant binding affinity of PI polyamides with DNA [12]. We herein assess the efficacy of PI polyamides, with 8-bp sequence recognition, that contain strategically placed discrete heterocycle/β pairs at the cellular level and demonstrate the potentials of such substitutions in 12-ring PI polyamides in vivo.
Syntheses of PI polyamides
PI polyamides were synthesized in a stepwise reaction based on a previously described Fmoc solid-phase protocol [5] using an automated solid-phase peptide synthesizer (PSSM-8, Shimadzu Industry) at 10 μmol scales (9.8 mg) of Fmoc-β-alanine Wang resin (NOVA Chemicals). After the synthesis, Dp (Wako) was mixed with the resin at 65ºC for 12 h for compound cleavage. Purification of PI polyamides was performed using high-performance liquid chromatography (LC-20, Shimadzu Industry), using a 10 mm×150 mm Phenomenex Gemini-NX3u 5-ODS-H reverse-phase column (Phenomenex) in 0.1% acetic acid in water with acetonitrile as eluent, at a flow rate of 10 ml/min, and a linear gradient from 0% to 66.7% acetonitrile over 20 min, with detection at 310 nm. Collected fractions were analyzed by LC-MS. Polyamide 1. m/z calculated for C82H96N32O15, [M+H]+ 1770.85; found 1770.65, [M+2H]2+ 886.4; found 885.90, [M+3H]3+ 591.28; found 591.10, [M+4H]4+ 443.71; found 443.50. Polyamide 2. m/z calculated for C79H95N31O15, [M+2H]2+ 860.40; found 860.35, [M+3H]3+ 573.93; found 574.05, [M+4H]4+ 430.70; found 430.80. Polyamide 3: LC-MS m/z calculated for C76H94N30O15, [M+2H]2+ 834.88; found 834.85, [M+3H]3+ 556.92; found 557.05, [M+4H]4+ 417.94; found 418.00. Polyamide 4: m/z calculated for C76H94N30O15, [M+H]+ 1668.75; found 1668.65, [M+2H]2+ 834.88; found 834.85, [M+3H]3+ 556.92; found 557.00, [M+4H]4+ 417.94; found 418.00. Polyamide 5: LC-MS m/z calculated for C76H94N30O15, [M+H]+ 1668.74; found 1668.29 (Figures 1S-5S).
Surface plasmon resonance (SPR) assay
All SPR experiments were performed on a ProteON XPR36 (Bio- Rad) at 25ºC as described previously [10]. Biotinylated hairpin MMP-9 nucleotides (5’biotin-GACCCCTGAGTCAGCACTTGCCTTTTGGCAAGTGCTGACTCAGGGGTC- 3’, Operon) and hairpin MYCN DNA (5’ biotin-TGGCTTTTGGCGCGAAAGTTTTCTTTCGCGCAAAACGGA- 3’, Operon) were immobilized on a streptavidin coated SA sensor chip at a flow rate of 20 μL/m to obtain the required immobilization level (up to approximately 1000 resonance units (RU)). Samples were dissolved in HBS-EP buffer (10 mM 4-(2-hydroxyethyl)- 1-piperazineethanesulfonic acid (HEPES), 150 mM NaCl, 3 mM ethyl-enediamine tetraacetic acid (EDTA), and 0.005% surfactant P20) with 0.1% DMSO at 25ºC, pH 7.4 for the experiment.
Cell culture, RT-PCR and Real-time PCR
MDA-MB-231 cells (ATCC) were cultured in DMEM (Life Technologies), and CHP134 cells (Roswell Park Cancer Institute, Buffalo, NY) were cultured in RPMI1640 (Life Technologies); both media were supplemented with 100 μg ⁄ mL streptomycin, 100 units ⁄ mL penicillin (Gibco), and 10% FBS (Gibco) in a humidified environment and 5% CO2 at 37ºC. The MDA-MB-231 cells were treated with PI polyamide (3 μM) with 0.1% DMSO and control cells were administered with 0.1% DMSO for 48 h. Similarly, CHP134 cells were treated with PI polyamide (5 μM) and control cells with 0.1% DMSO for 72 h. Cell fractions were processed for RNA isolation with RNAeasy plus mini kit (Qiagen) followed by first-strand cDNA synthesis with SuperScript VILO cDNA Synthesis System (Thermo Fisher) according to the manufacturer’s instructions. Real-time PCR and subsequent calculations were performed with Power SYBR Green Master Mix reagents (Applied Biosystems) on a 7500 Real-Time PCR System (Applied Biosystems). The PCR mixture contained 1x Master Mix, 200 nM forward and reverse primers, 1.0 μL of the first strand cDNA mix, and 0.25 μM AmpErase uracyl N-glycosylase (UNG) in a total volume of 20 μL. To permit UNG cleavage for 2 min at 50ºC, the PCR enzymes were activated by 10 min incubation at 95ºC. Each of the 40 PCR cycles consisted of a 15 s denaturation step at 95ºC and a hybridization-elongation step for 1 min at 60ºC. The standard curve was obtained using 5 volumes of serial dilutions of pooled first strand cDNA from 0.1% DMSO-treated control cells. The primers for MMP-9 (forward, 5’-GAGACCGGTGAGCTGGATAG-3’; reverse, 5’-TACACGCGAGTGAAGGTGAG-3’), for MYCN (forward, 5’-CTTCTTAGCTGCACCAGTGAC-3’; reverse, 5’-AGTGATGGTGAATGTGGT-3’) and for GAPDH as the internal control (forward, 5’-GCACCGTCAAGGCTGAGAAC-3’; reverse, 5’-TGGTGAAGACGCCAGTGGA-3’) were used. The relative quantity for the MMP-9 or MYCN genes was normalized for GAPDH-expression.
Among the candidate PI polyamides 1, 2, 3, and 4 (Figure 1), compound 1 was designed to be a structurally rigid polyamide in which two hexameric heterocycle units are joined by a γ-aminobutyric acid (γ-turn) moiety. A β subunit was then introduced to the scaffolds in 2, 3, and 4 (“β-polyamides”) to assess improvements in molecular flexibility. Polyamide 2 contained a single β subunit at the 3/3’ position as a Py/β pair (β1- polyamide), while 3 and 4 (β2-polyamide) contain two βs in their main scaffolds. In compound 3, the two βs are located at the 3/3’ position as a β/β matched pair, whereas in 4 the two β units are configured as discrete pairs (Py/β and β/Im) at the 3/3’ and 4/4’ positions, respectively (Figure 1). The PI polyamides 1, 2, 3, and 4 were synthesized by the Fmoc solid-phase synthesis, with the Wang resin as the solid support [13]. Stepwise oligomerization of Py, Im, γ-turn, and β for the main scaffold was achieved using HCTU as the coupling reagent. Simultaneous cleavage from the resin and end-capping with the Dp tail was conducted as previously described [10] to afford final compounds 1-4. Compound purities were confirmed by reverse-phase HPLC. Surface plasmon resonance (SPR) analyses were conducted to evaluate the binding affinities of these polyamides to the target DNA 5’-AGTCAGCA-3’ sequence (Figure 2 and Table 1). Compounds 2, 3, and 4 displayed appreciable SPR responses while polyamide 1 was unable to produce a detectable signal. Such responses suggested that rigid structures in absence of β subunits were likely incapable of binding the minor groove at similar affinities. Dissociation equilibrium constants (KD=kd/ka, in which kd and ka were the dissociation and association rate constants, respectively) between the free and bound states of these polyamides were determined as an exponential decay model (Figure 2) [14]. Polyamides 3 (4.33 ± 0.51×10–8 M) and 4 (5.00 ± 0.94×10–8 M) were two orders of magnitude lower than 2 (123 ± 16×10–8 M, Table 1) in KD, indicating that compound 2 had a lower affinity for 5’-AGTCAGCA-3’ compared to 3 and 4. This result also inferred that β2- polyamides were potentially more flexible than β1-polyamides due to the absence of a rigid hexameric heterocyclic chain. Both association (ka) and dissociation (kd) rates were higher for β2-polyamides 3 and 4 compared to β1-polyamide 2, further indicating that higher structural flexibility afforded by the additional the β substitution potentially accelerated not only the association but also the dissociation (Table 1). The accelerated dissociation would ascribe to a decrease of interaction associated with the π-system like CH-π and π-π interactions as a result of lowering the number of heterocycles by substitutions to aliphatic β-alanines. We previously reported that polyamide 3 inhibited the expression of MMP-9 mRNA in MDA-MB-231 cells by binding to the 5’-AGTCAGCA-3’ sequence at the AP-1 binding site [10]. To investigate the effect of quantity and positioning of β substitutions in PI polyamide structure on gene-silencing, we again evaluated MMP-9 mRNA expression levels in MDA-MB-231 cells treated with compounds 1-4 (3 μM with 0.1% DMSO, 48 h) and 0.1% DMSO as the control (Figure 3). Polyamides 2 (86%), 3 (50%), and 4 (49%) showed distinct mRNA silencing in MDA-MB-231 cells, in sharp contrast to compound 1.
Figure 2: SPR sensorgrams for the interaction of (a) 1, (b) 2, (c) 3 and (d) 4 with hairpin DNAs containing the sequence 5’-AGTCAGCA-3’ immobilized on the surface of a sensor chip SA. Each five curves of the lowest, mid low, middle, mid high and highest indicates concentrations of the PI polyamides 25, 50, 100, 200 and 400 nM, respectively. All the experiments were performed in HBS-EP buffer (10 mM HEPES, pH 7, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) with 0.7% DMSO (v/v) at 25 ºC.
| PI polyamide | KD [10-8M]a | ka[103 M-1s-1]b | kd [10-8s-1]c | |
|---|---|---|---|---|
| 1 | 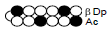 |
N.D | N.D | N.D |
| 2 | 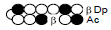 |
123±16 | 5.19±0.13 | 6.37±0.83 |
| 3 | 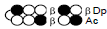 |
4.33±0.51 | 254±51 | 11.0±2.7 |
| 4 | 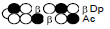 |
5.00±0.94 | 474±0.90 | 23.7±0.16 |
All values are evaluated from at least five times SPR experiments with standard deviations.
aDissociation Constant, bAssociation rate constant, cDissociation rate constant
Table 1: Binding affinities of the PI polyamides 1, 2, 3, and 4 with the
sequence 5’-AGTCAGCA-3’.
Inhibition activities strongly correlated with KD values determined from SPR in Table 1, indicating that β-polyamides were capable of binding the intended 5’-AGTCAGCA-3’ nucleotide sequences in vitro. The fact that we observed comparable activities from both compounds 3 and 4 was particular noteworthy, as it implied that the use of Py/β-β/ Im pairs could potentially expand the range of nucleotide recognition, without loss in PI polyamide and DNA binding affinity, which is difficult to achieve with β/β-polyamides alone.
We thus expanded the target binding site to a repeating GC sequence, 5’-TTGGCGC-3, at the E2F binding site within MYCN promoter of MYCN amplified CHP134 human neuroblastoma cells to explore the Im/β-β/Im polyamides. We synthesized β2-polyamides 5 with two discrete Im/β and β/Im pairs that recognizes the G/C and C/G base pairs at the 3/3’ and 4/4’ positions, respectively (Figure 4a). Polyamide 5, with KD=3.07 ± 0.67×10-8 M from SPR, also suggested binding with the intended 5’-TTGGCGC-3’ sequence, with the binding affinity sufficiently enough to suppress protein-DNA interactions (Figure 5). Compound 5 had a KD value comparable to those of 3 and 4, suggesting that their relative affinities to the target sequences were on the same order. Indeed, mRNA expressions in CHP134 cells treated with 5 also significantly decreased (65%) compared to untreated cells, indicating that 5 indeed could penetrate CHP134 cells and, upon binding its target sequence, suppress transcription factor binding (Figure 4b). These findings provided a rationale for designing PI polyamides with two β substitutions, in an adjacent Im/β and β/Im configuration, as a promising strategy to targeting 8-bp or longer DNA sequences, particularly in G/C rich regions.
Figure 4: PI polyamide 5 targeting 5’-TTGGCGC-3 in the MYCN promoter. (a) chemical structure of 5. (b) MYCN mRNA expression level in CHP134 cells treated with 5 (5 μM, 72 h) relative to the control. Error bars indicate mean ± SD (n=3 for each group). *p<0.01 by unpaired Student’s t-test compared to DMSO.
Figure 5: SPR sensorgrams for the interaction of 5 with hairpin DNAs containing the sequence 5’-TTGGCGCGA-3’ immobilized on the surface of a sensor chip SA. Each five curves of the lowest, mid low, middle, mid high and highest indicates concentrations of the PI polyamides, 25, 50, 100, 200, and 400 nM, respectively. All the experiments were performed in HBS-EP buffer (10 mM HEPES, pH 7, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) with 7.0% DMSO (v/v) at 25ºC. KD: 3.07 ± 0.67[10-8M], ka: 1.76 ± 0.34[104M-1s-1], kd: 5.19 ± 0.42[10-4s-1].
We hereby demonstrated an efficient molecular design of 12-ring PI polyamides targeting 8-bp sequences at the cellular level. Polyamides with two discrete heterocycle/β pairs were superior in DNA binding affinity and the range of target sequence recognition. Gene-silencing experiments with the Py/β-β/Im-polyamide rationally designed to target a GC-rich motif (5’-TTGGCGC-3) in the MYCN promoter further demonstrated the inhibitory activity of the polyamide in mRNA expressions.
This work was supported by the KIBAN-B fund (26290060) from MEXT, Japan (TW, KS and HN), the Takeda Science Foundation (KS), the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT) (TW, KS and HN), and Inohana-Shougakukai (TW).