Journal of Developing Drugs
Open Access
ISSN: 2329-6631
ISSN: 2329-6631
Research Article - (2022)Volume 11, Issue 2
Array technology; Pharmacogenomics; Combinatorial chemistry; Proteomics; New Chemical Entities (NCE)
The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of small molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is often referred to as computer-aided drug design [1]. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. Drug Design molecules that bind to a target (e.g. protein, nucleic acid). It relies on prior knowledge of the structure, function, and mechanism of the target, thereby avoiding random testing of thousands of molecules.
Selected/designed molecule should be
• Organic small molecule
• Complementary in shape to the target
• Oppositely charge to the bio-molecular target
The molecule will
• Interact with target
• Bind to the target
• Activates or inhibits the function of a bio-molecule such as a protein
Novel drug approaches like
• Computer Aided Drug Design (CADD)
• Molecular modeling
• Structure based drug design
• Analog drug design
• Combinatorial chemistry
• Computational chemistry
• Array technology
• Pharmacogenomics
• Combinatorial Chemistry
• Proteomics
• Recombinant DNA technology etc.
• Array technology
• Pharmacogenomics
• Combinatorial chemistry
• Proteomics
• Recombinant DNA technology.
Array technology
Based on RNA and DNA hybridization reaction:
• Simultaneously several of genes can be analyzed.
Advantages:
• Miniaturization.
• Development of fluorescent labeled nucleotides, which is detected by laser screening.
• Decreased use of hazardous and cumbersome radioactive labeled DNA.
Methods used:
• Chips located with synthetic oligonucleotides.
• Chips located with a DNA fragment isolated directly from respective genes (Figure 1).
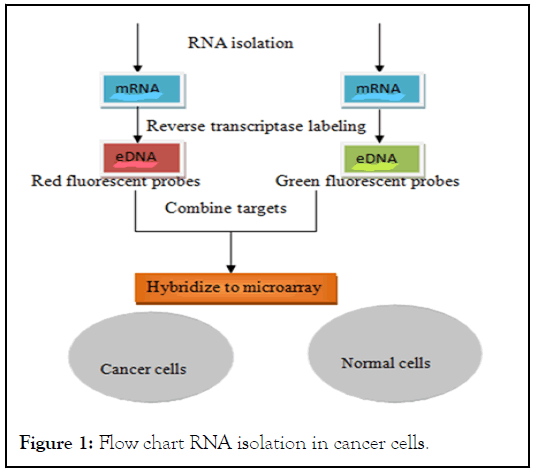
Figure 1: Flow chart RNA isolation in cancer cells.
Types:
• DNA microarray
• RNA microarray
• Protein microarray
• Antibody microarray
• Tissue microarray
Application of microarray:
• Study regulatory networks
• Drug discovery-mechanism of action
• Diagnostics-tumor diagnosis
Genomic DNA hybridizations:
• Explore microbial diversity
• Whole genomic comparisons-genome evolution
• Identify DNA biding sites
• Diagnostics-tumor diagnosis
Pharmacogenomics
Pharmacogenomics is the study of the role of the genome in drug response. Its name reflects it’s combining of pharmacology and genomics. Pharmacogenomics analyzes how the genetic makeup of an individual affects his/her response to drugs [2]. Pharmacogenomics was first recognized by Pythagoras around 510 BC when he made a connection between the dangers of fava bean ingestion with hemolytic anemia and oxidative stress [3]. This identification was later validated and attributed to deficiency of G6PD in the 1950s and called favism.
There are several known genes which are largely responsible for variances in drug metabolism and response. The focus of this article will remain on the genes that are more widely accepted and utilized clinically for brevity (Figure 2).
Figure 2: Cancer Pharmacogenomics.
• Cytochrome P450s
• VKORC1
• TPMT
Pharmacogenomics aims to develop rational means to optimize drug therapy, with respect to the patients' genotype, to ensure maximum efficiency with minimal adverse effects. Through the utilization of pharmacogenomics, it is hoped that pharmaceutical drug treatments can deviate from what is dubbed as the "one-dose-fits-all" approach. Pharmacogenomics also attempts to eliminate the trial-and-error method of prescribing, allowing physicians to take into consideration their patient's genes, the functionality of these genes, and how this may affect the efficacy of the patient's current or future treatments [4].
Application:
The list below provides a few more commonly known applications of pharmacogenomics:
• Improve drug safety, and reduce Adverse drug reaction (ADRs)
• Tailor treatments to meet patients' unique genetic predisposition, identifying optimal dosing
• Improve drug discovery targeted to human disease
• Improve proof of principle for efficacy trials.
Pharmacogenomics may be applied to several areas of medicine, including pain management, cardiology, oncology, and psychiatry. A place may also exist in forensic pathology, in which pharmacogenomics can be used to determine the cause of death in drug-related deaths where no findings emerge using autopsy. And, used in cancer treatment, cardiovascular disorders, psychiatry disorders, etc.
Example case studies:
• Case A: Antipsychotic adverse reaction
• Case B: Pain management
• Case C: FDA warning on codeine overdose for infants
Proteomics
Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions. The proteome is the entire set of proteins that is produced or modified by an organism or system. Proteomics has enabled the identification of ever increasing numbers of protein. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain that has benefitted greatly from the genetic information of various genome projects, including the Human Genome Project. It covers the exploration of proteomes from the overall level of protein composition, structure, and activity. It is an important component of functional genomics [5]. Proteomics generally refers to the large-scale experimental analysis of proteins and proteomes, but often is used specifically to refer to protein purification and mass spectrometry.
Proteomics is the study of proteomes, which are the collections of proteins expressed in cells. Whereas genomes are essentially invariant in different cells in an organism, proteomes vary from cell to cell, with time and as a function of environmental stimuli and stress [6,7].
The first studies of proteins that could be regarded as proteomics began in 1975, after the introduction of the twodimensional gel and mapping of the proteins from the bacterium Escherichia coli.
Methods
Three methods for separation of complex protein or peptide samples are preferred in proteomics: (Figure 3).
Figure 3: Flow chart of Proteomic analysis.
• Denaturing Polyacrylamide Gel Electrophoresis (PAGE) or Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
• Two-dimensional gel electrophoresis
• High-Performance Liquid Chromatography (HPLC)
Application:
• New Drug Discovery
• Interaction proteomics and protein networks
• Expression proteomics
• Biomarkers
• Proteogenomics
• Structural proteomics
Trends:
A number of emerging concepts have the potential to improve current features of proteomics. Obtaining absolute quantification of proteins and monitoring post-translational modifications are the two tasks that impact the understanding of protein function in healthy and diseased cells. For many cellular events, the protein concentrations do not change; rather, their function is modulated by Post-Translational Modifications (PTM).
• System biology
• Human plasma proteome
The various approaches used in drug design include the following
• Random screening of synthetic compounds or chemicals and natural products by bioassay procedures.
• Preparation of structural analogs of lead with increasing biological activity.
• Application of bio isosteric principle.
The current trend in the drug design is to develop new clinically effective agents through the structural modification of lead nucleus. The lead is a prototype compound that has the desired biological or pharmacological activity but may have many undesirable characteristics, like high toxicity, other biological activity, and insolubility or metabolism problems. Such organic leads once identified, are easy to exploit. This process is rather straightforward. The real test resides with the identification of such lead real test resides with the identification of such lead bioactive positions on the basic skeleton of such leads.
The rate of drug design and discovery is dependent on the ability to identify, characterize novel, patentable newer target drug molecules, usually termed as New Chemical Entities (NCE’s), which is essentially possess the inherent capability and control of a specific disease/ailment. Besides, being efficacious and safer in character. With the advent of latest technological advancements in the specialized areas related to genomics and combinatorial chemistry an appreciable in the R&D strategies. NCE status, position and recognition is an absolute must not only to ensure marketing exclusively but also to justify the huge investment in the ensuring R&D process there by making more or less core element of the entire drug design and discovery process (Figure 4).
Figure 4: New techniques and strategies in drug discovery.
New techniques and strategies in drug design and discovery
• Quantum chemistry
• Virtual target profiling
• In silico autophagy methods
• Histone deacetylases
• Allosteric kinase inhibitors
• Innovative antiviral agents
• Nitric oxide-based anticancer agents
• Photoactivation strategies
Novel approaches in drug design are the creative process of finding new remedies based on the knowledge of a biological target. This review discusses principle of drug design, various approaches of drug design, lead discovery, lead modification and various types of drug discovery. Bio-isosteres is an important lead modification approach that has been shown to be useful to attenuate toxicity or to modify the activity of a lead, and may have a significant role in the alteration of pharmacokinetics of a lead. The process of drug discovery by laboratory experiments is time consuming and very expensive as compared to computational methods and play an vital role in Pharmaceutical Development of new drug for therapeutic efficacy.
[Crossref] [Google scholar] [Pubmed]
[Crossref] [Google scholar] [Pubmed]
[Crossref] [Google scholar] [Pubmed]
[Crossref] [Google scholar] [Pubmed]
[Crossref] [Google scholar] [Pubmed]
[Crossref] [Google scholar] [Pubmed]
Citation: Senthil S, Malathi T, Surendrakumar M (2022) Drug Design and its Novel Approaches. J Develop Drugs. 11:166.
Received: 10-Mar-2022, Manuscript No. EOED-22-11187; Editor assigned: 14-Mar-2022, Pre QC No. EOED-22-11187 (PQ); Reviewed: 28-Mar-2022, QC No. EOED-22-11187; Revised: 04-Apr-2022, Manuscript No. EOED-22-11187 (R); Published: 11-Apr-2022 , DOI: 10.35248/2329-6631.22.11.166
Copyright: © 2022 Senthil S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.