Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2021)Volume 14, Issue 3
Coronavirus pandemic COVID 19 has caused a wide range of harm worldwide with its inception in December 2019 in Wuhan, China. Till date there is no promising drug identified for the treatment of disease. In the view of this, scientists have elucidated X-ray structures of the proteins in SARS-CoV-2 virus. These can act as probable drug targets for the designing of drugs which is urgent need. One of the main proteins of the virus is its main protease Mpro which is responsible for producing polyproteins of the virus. In this study we have used main protease as the target for drug design and repurposing for COVID-19. Two approaches were applied in order to develop a fast and effective treatment against the virus. Drug repurposing through in-silico docking analysis of existing FDA approved drugs was one method and high throughput screening of molecules from the ZINC database against main protease was the other technique applied. Two docking protocols were utilized- a fast docking algorithm to screen the hits or lead molecules followed by simulation based molecular dynamics docking procedure to optimize the obtained hits. We could observe a definite scaffold based binding affinity against the main protease. These scaffolds were lutein, steroids, morphine and quinolone, CPT. Thiotepa was identified as the best docked molecule with highest binding affinity. Unique molecules like lutein, beta carotene, Buprenorphine etc. were identified which can be used as repurposed drugs against SARS-CoV-2. Also these scaffolds show unique pharmacophores which can be utilized to design potential novel leads against SARS-CoV-2 for future treatment.
Drug repurposing; SARS-CoV-2; Main protease; HTVS; Simulation; Docking
Coronavirus sickness 2019 (COVID-19) is a transmittable malady brought about by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) an encompassed, positive-sense, single-stranded RNA beta coronavirus of the family Coronaviridae [1]. The infection was first recognized in December 2019 in Wuhan, China, and has spread all around from that point forward, bringing about the progressing 2019-20 coronavirus pandemic [2]. Starting at 24 April 2020, more than 2.79 million cases have been affirmed across 185 nations and regions, causing in excess of 195,000 passings. In excess of 781,000 individuals have recouped [3].
In comparison with MERS or SARS, SARS-CoV-2 seems to spread all the more effectively, making it hard to contain and thus expanding its pandemic potential. To devise therapeutic procedures to balance SARS-CoV-2 contamination, it is critical to build up a far reaching comprehension of how this coronavirus captures the host over the span of disease, and to apply this information towards creating both new medications and repurposing existing ones. Up until this point, no clinically accessible antiviral medications have been created for SARSCoV, SARS-CoV-2 or MERS-CoV [4]. Clinical trials are continuously ongoing for treatment of COVID-19 with the nucleotide simple RNA-subordinate RNA Polymerase (RdRP) inhibitor remdesivir, and ongoing information recommends another nucleotide analogs might be successful against SARSCoV- 2 contamination in research facility animals.
The virions of SARS-CoV-2 comprise of crown-formed peplomers, 80-160 nm in diameter, and a ~30 kb long singlestranded RNA particle of positive extremity with 5' top and 3' Poly-A tail [5]. The RNA genome is made out of in any event six open reading frames (ORFs) of which the first ORF encodes two polypeptides pp1a and pp1ab the two of which besides prompts the creation of 16 nonstructural proteins (nsPs). Different ORFs that make up the staying 33% of the viral genome offer ascent to the creation of four primary structural proteins of the virion: Spike protein (S), Envelope protein (E), Membrane protein (M) and Nucleocapsid protein (N) [6]. One of the best-described medication focuses among coronaviruses is the principle protease (Mpro, likewise called 3CLpro). Alongside the papainlike protease(s), this protein is basic for handling the polyproteins that are deciphered from the viral RNA. The Mpro works at no less than 11 cleavage destinations on the huge polyprotein 1ab (replicase 1ab, ~790 kDa); the acknowledgment grouping all things considered locales is Leu-Gln ↓ (Ser, Ala, Gly) (↓ marks the cleavage site). Hindering the action of this compound would hamper the replication. Since no human proteases with comparative cleavage explicitness are known, such inhibitors are probably not going to be poisonous.
Drug repositioning (also called drug repurposing) involves the investigation of existing drugs for new therapeutic purposes. In recent years drug repurposing screens have emerged as a resourceful alternative to fasten the drug development process against rapidly spreading emerging infections. This same technique has also been deployed in this work against the SARSCoV- 2. The existing drugs approved by FDA have been taken into consideration for this study so as to identify potential drugs that can act against SARS-CoV-2. Also we have undertaken another strategy to identify lead candidates from already existing databases by virtually screening them and identifying scaffolds that show activity against SARS-CoV-2.
The main protease enzyme was used as a potential target for the docking and screening of library compounds. The threedimensional structure is highly similar to that of the SARS-CoV Mpro, with 96% sequence identity. Domains I and II (aa 10 to 99 and 100 to 182, separately) are six- stranded antiparallel β barrels that harbor the substrate-restricting site between them. Domain III (aa 198 to 303), a globular group of five helices, is associated with directing the dimerization of the Mpro, chiefly through a salt-connect between Glu290 of one protomer and Arg4 of the other [7]. The recently elucidated crystal structure of Main protease from SARS-CoV-2 with PDB id 6LU7 was used for the study (Figure 1).
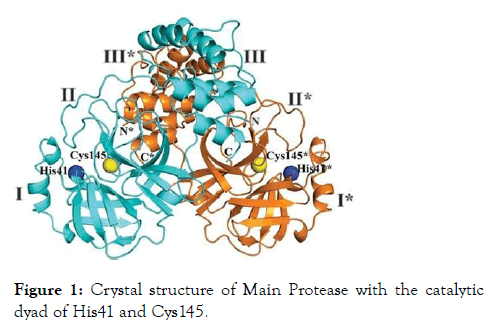
Figure 1: Crystal structure of Main Protease with the catalytic dyad of His41 and Cys145.
Docking analysis of receptor-ligand interactions
The receptor active site cavity was identified by already existing ligand in the crystal structure complex. A two-step docking protocol was implemented- High throughput virtual screening algorithm that identifies hotspots (Libdock) followed by CHARMm based molecular dynamics scheme (CDocker) to dock ligands into the active site. The Libdock gives a positive binding affinity score; the top poses from the docking results were shortlisted for CDocker based docking. The CDocker gives a negative binding affinity score and the molecules having least binding affinity were selected as leads and potential drugs for repurposing that can act against SARS-CoV-2 [8].
The Libdock docking program performs the following steps using a set of pre-generated ligand conformations and a receptor with a specified binding site: Libdock uses protein site features referred to as HotSpots. HotSpots consist of two types: polar and apolar. A polar Hotspot is preferred by a polar ligand atom (for example a hydrogen bond donor or acceptor) and an apolar HotSpot is preferred by an apolar atom (for example a carbon atom). The receptor HotSpot file is calculated prior to the docking procedure. The rigid ligand poses are placed into the active site and HotSpots are matched as triplets. The poses are pruned and a final optimization step is performed before the poses are scored. Ligand hydrogens, which are removed during the docking process, are added to the ligand poses. These hydrogens are not optimized, so they require further optimization to ensure that receptor-ligand hydrogen bonds are formed correctly.
The high throughput screening of ligands was done using the FAST method of conformation. Energy threshold was set at 20 and maximum conformations were set at default value of 255. The number of hotspots was at the default value of 100. The top poses of each ligand were shortlisted. After selecting the top poses from Libdock the molecules were subjected to CDocker docking program [9].
CDOCKER is a grid-based molecular docking method that employs CHARMm. The receptor is held rigid while the ligands are allowed to flex during the refinement. For predocked ligands, prior knowledge of the binding site is not required. Random ligand conformations are generated from the initial ligand structure through high temperature molecular dynamics, followed by random rotations. The random conformations are refined by grid-based (GRID 1) simulated annealing and a final grid-based or full force field minimization. The following steps are included in the CDOCKER protocol:
1. A set of ligand conformations are generated using hightemperature molecular dynamics with different random seeds.
2. Random orientations of the conformations are produced by translating the center of the ligand to a specified location within the receptor active site, and performing a series of random rotations. A softened energy is calculated and the orientation is kept if the energy is less than a specified threshold. This process continues until either the desired number of low-energy orientations is found, or the maximum numbers of bad orientations have been tried.
3. Each orientation is subjected to simulated annealing molecular dynamics. The temperature is heated up to a high temperature then cooled to the target temperature.
4. A final minimization of the ligand in the rigid receptor using non-softened potential is performed.
5. For each final pose, the CHARMm energy (interaction energy plus ligand strain) and the interaction energy alone are calculated. The poses are sorted by CHARMm energy and the top scoring (most negative, thus favorable to binding) poses are retained.
The simulation docking algorithm used the default parameters of generating top 10 hits. The dynamic steps were set at 1000 and the dynamic temperature was also considered to be 1000. The maximum orientations to refine were 10. The simulated annealing model was chosen for simulation procedure with heating steps set at 2000, heating target temp of 700, number of cooling steps were 5000 and cooling target temperature was 300. After CDocker the top poses having lowest binding affinity and best receptor-ligand interactionsare chosen as best molecules those have the potential to act against COVID-19.
The active site cavity of the Main protease enzyme was identified based on the crystal structure of the protein. The active site was identified around the existing inhibitor with sphere radius of 13.807549 and XYZ coordinates were -10.7118, 12.4113 and 68.8312 respectively.
The active site residues include amino acids from 24-26, 41, 49, 54, 140-145 and 163-192. The following Figure 2 shows the active site sphere of the main protease.
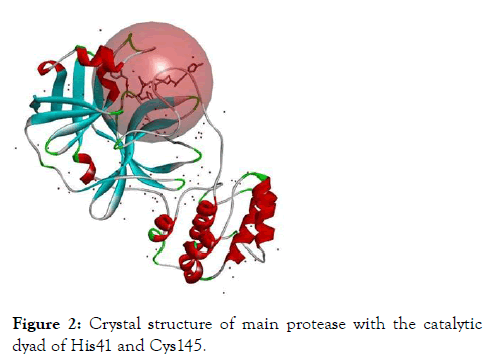
Figure 2: Crystal structure of main protease with the catalytic dyad of His41 and Cys145.
Out of the 3600 FDA approved drugs used for docking in Libdock for the repurposing against Covid-19, top 2978 molecules were shortlisted for CDocker analysis. Out of the 314 molecules from the ZINC databases 249 molecules were shortlisted for CDocker analysis. A total of 3227 molecules were used for CDocker study.
Out of the 2978 molecules subjected to CDocker analysis, top 700 molecules were shortlisted based on the score and binding affinity value. The 700 molecules were classified based on their scaffold specific activity. The best scaffolds identified by simulation based docking were lutein, steroid, morphine, quinolone, CPT and cephalosporin scaffold. Thiotepa was identified as the best docked drug against the SARS-CoV-2 protease. From the individual scaffolds lutein showed highest binding affinity followed by fusidic acid. Pharmacophoric mapping of these scaffolds revealed specific interactions with the active site residues which can facilitate novel drug design of analogs based on these scaffolds. The best docked drug Thiotepa showed a binding affinity of -245.519.
The next best docked molecule was the fusidic acid which resembles cholestan scaffold molecule, with the binding affinity of -91.3714. It is an antibiotic with established antibacterial activity. The next best molecule was lutein. Its binding energy was -84.3549. Most of its analogs were found in the top 30 molecules, thus establishing lutein scaffold to act on the protease protein. Other molecules that showed better binding were Rocuronium bromide and beta carotene which again belong to the cholestan family, with binding affinity -63.6734 and -76.6497 respectively.
One of the important scaffolds that were identified was of the camptothecin molecule.Topotecan molecule showed best binding affinity of -69.8488 and Libdock score of 145.016. Ajmaline a plant alkaloid was also found to show good binding affinity towards the main protease with binding affinity of -77.8805.
Morphine scaffold was also found to be active against the main protease. 2 molecules Buprenorphine and Nalbuphine were found to show good affinity with docking score of -67.4387 and -62.0371 respectively. Also one of the important scaffolds that were recognized as having good binding affinity was Quinolone scaffold. The molecules like Gatifloxacin and Grepafloxacin showed a docking score of -46.9551 and -45.6419 respectively. Beclomethasone, Paramethasone, Betamethasone were some of the other steroids that were identified showing good binding affinity of -46.9818, -45.5, -44.7844 respectively.
Table 1 above shows the top 30 molecules shortlisted based on docking score and binding interactions from the existing FDA database. Thus mostly all of the top 30 molecules belong to cholestans, lutein, camptothecin, morphines and quinolone scaffolds, with exception of a few drugs that were different. All these molecules show a distinct pharmacophore based on which novel leads against Covid-19 can be developed and designed.
| Molecule | Molecule Name | Cdocker Score | Libdock Score |
|---|---|---|---|
| ZINC01530867 | Thiotepa | -245.519 | 69.4739 |
| ZINC14243816 | Fusidic acid | -91.3714 | 122.439 |
| ZINC14879959 | Lutein | -84.3549 | 113.997 |
| ZINC08220548 | Ajmaline | -77.8805 | 100.21 |
| ZINC06845076 | Beta-Carotene | -76.6497 | 106.593 |
| ZINC35018256 | Topotecan | -69.8488 | 145.016 |
| ZINC38321402 | Buprenorphine | -67.4387 | 121.502 |
| ZINC53229445 | Rocuronium bromide | -63.6734 | 109.413 |
| ZINC03977862 | Nalbuphine | -62.0371 | 116.755 |
| ZINC29051126 | Retapamulin | -59.2069 | 138.768 |
| ZINC11616424 | Tibolone | -59.1548 | 113.45 |
| ZINC03830615 | Cyproterone | -56.5033 | 110.924 |
| ZINC02039583 | Isocodeine HCl | -55.6457 | 98.2887 |
| ZINC33938184 | Calcipotriolhyd rate | -54.7226 | 135.549 |
| ZINC03831458 | B-sitosterol | -52.4574 | 126.077 |
| ZINC04097416 | Desogestrel | -51.8337 | 102.095 |
| ZINC03830441 | Cefotetan | -51.4352 | 112.046 |
| ZINC03881949 | Norethynodrel | -49.2817 | 112.084 |
| ZINC03830226 | 5-androstenediol | -49.1489 | 94.279 |
| ZINC03831095 | Methylpredniso lone acetate |
-49.0357 | 115.474 |
| ZINC03927200 | Drospirenone | -48.2456 | 115.429 |
| ZINC04097285 | Beclomethasone | -46.9818 | 105.501 |
| ZINC38197764 | Gatifloxacin | -46.9551 | 117.005 |
| ZINC04097404 | Vecuronium | -46.5503 | 118.917 |
| ZINC03977777 | Amcinonide | -46.4805 | 118.309 |
| ZINC00967787 | Grepafloxacin | -45.6419 | 125.04 |
| ZINC03831268 | Paramethasone | -45.5 | 105.835 |
| ZINC03830287 | Betamethasone | -44.7844 | 109.953 |
| ZINC03814394 | Gestodene | -44.543 | 106.403 |
Table 1: Molecular docking analysis of selected compounds
SARS-CoV-2 a highly infectious and pathogenic coronavirus has become global concern. The lack of availability of approved treatment for this disease calls forth the scientific community to find novel compounds with the ability to treat it. This study used FDA database for drug repurposing and other databases for design and development of novel molecules. Our study proposes drugs that can be used immediately against SARS-CoV-2 identified through drug repurposing, also at the same time proposes receptor based pharmacophotic maps distinct for each scaffold that can be used to develop and design novel molecules against COVID-19 in future. Thus molecules like the Thiotepa, Lutein, Buprenorphine, Gatifloxacin, Beclomethasone, Ajmaline and Topothecan can act as potential drugs against SARS-CoV-2 with main protease as their target. These results can be taken up for further in vitro and in vivo trials.
Authors reports no conflict of interest.
On behalf of Amity Institute of Biotechnology, Amity University, Mumbai we thank Dassaults Systems for sponsoring BIOVIA licence to support the challenging COVID-19 drug research.
Citation: Bastikar VA, Gupta P, Sangshetti JN, Bastikar AV, Chhajed SS (2021) Drug Repurposing Approach Targeting Main Protease Using HTVS and Pharmacophoric Mapping: Exceptional Reassuring Itinerary for Most Insolvent Anti-SARS-CoV-2 Drug. J Proteomics Bioinform. 14:527.
Received: 18-Feb-2021 Accepted: 04-Mar-2021 Published: 11-Mar-2021 , DOI: 10.35248/0974-276X.21.14.527
Copyright: © 2021 Bastikar VA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : None