Journal of Food: Microbiology, Safety & Hygiene
Open Access
ISSN: 2476-2059
ISSN: 2476-2059
Research Article - (2020)Volume 5, Issue 1
Representative grains of Nigerian local (Igbemo) rice obtained from Akure, Nigeria were cooked and fermented under aseptic conditions for seven days. Fungi and bacteria associated with the sample before and during the fermentation were isolated and characterized. The pH and sensory attributes of the rice samples were also determined as fermentation progressed. Among the diverse bacteria isolates obtained from the food, only Bacillus cereus and B. subtilis were isolated consistently with populations that ranged between 1.0 × 103-1.9 × 104 cfu/g. Others were B. megaterium, B. maceans, B. polymyxa, B. brevis, Erwinia amylovora and E. tracheiphila. The following fungi isolates were obtained from the rice before and during fermentation: Vigaria nigra, Varicoporium elodeae, Articulospora inflata, Penicillin italicum and Saccharomyces cerevisiae. While the first three were only isolated within the first day of fermentation, the fourth (P. italicum) with population range of between 2.0 × 103-3.0 × 103 sfu/g occurred till the third fermentation day. Saccharomyces cerevisiae (5.0 × 103-2.3 × 104 sfu/g) was however isolated on the second to seventh fermentation days. Also, as the fermentation progressed, pH of the rice sample decreased but increased on the seventh fermentation day. There were no noticeable improvements in the appearance and aroma of the rice as fermentation progressed. The fermented rice was most palatable and generally acceptable on the seventh day of fermentation, thus making it ideal for consumption at the stage. Further research need to be conducted to determine the food’s nutritive status as fermentation progresses.
Igbemo rice; Fermentation; Fungi; Bacteria; pH; Sensory
Rice is a food of great importance all over the world. The crop is the seed of the monocot plant Oryza . There are two species of domesticated rice- O. sativa and O. glaberrima . Oryza sativa is the most common and often cultivated rice variety in Africa, America, Australia, China, New Guinea and South Asia. Oryza glaberrima is domesticated in West Africa [1].
Rice as a cereal grain is the most important staple food for a large part (half) of the world's human population, especially in East, South, Southeast Asia, the Middle East, Latin America, and the West Indies. Populations in these areas subsist wholly or partially on the crop. Rice is the grain with the second highest worldwide production, after maize [2,3].
In sub-Sahara Africa, West Africa is the leading producer and consumer of rice [4]. The region accounts for 64.2% and 61.9% of total rice production and consumption respectively in Sub-Saharan Africa. Except for Burkina Faso and Niger, rice is a staple crop throughout West Africa, especially in Côte d'Ivoire, the Gambia, Guinea, Guinea Bissau, Liberia, Senegal, Sierra Leone and Nigeria [5].
In some parts of the globe, rice is not only widely consumed, but also made into different products which have been found to be beneficial to mankind [6]. Like other crops and fruits in sub-Sahara Africa, rice, particularly the locally produced ones are faced with the risks of glut which usually lead to mass wastage of the commodity in the growing/harvesting seasons. This among others may be as a result of lack of the knowledge of ways of transforming the food into diverse products and/or by-products beneficial to man and animals.
It has been observed over the years that in Nigeria and most West African countries, fermentation is being employed even by the locals on foods like cassava and maize as they are processed into several products, thus making their cultivation more profitable to the farmers and also helping in food preservation [7].
Studies on fermentation of foods have shown the inherent benefits which include the provision and preservation of vast quantities of nutritious foods in a wide diversity of flavour, aroma and texture and also nutritionally enrich the human diets. It is however pertinent to note that foods can be ‘made’ or ‘marred’ by microbial fermentations. Improper fermentation of foods can lead to spoilage [7].
In a nut shell, fermenting rice is good wisdom towards maximally utilizing the crop and at the same time enhancing its nutritional and health benefits.
This study is therefore aimed at determining the varieties and populations of microorganisms associated with the fermentation of Nigerian local rice known as Igbemo and the effects of their proliferation on the sensory attributes and degree of acidity and/or alkalinity of the food.
Microbiological analysis
Diluent (saline solution) and media [Nutrient Agar (NA), deMan Rogosa Sharpe Agar (MRS) and Potato Dextrose Agar (PDA)] were prepared according to manufacturer’s specification for isolation. Isolated microorganisms were characterized and identified using Bergery’s manual sas described by Babatuyi et al.
Determination of pH
The pH determination was carried out in accordance to the standard scientific method described according to the method of A.O.A.C. (2012). The pH value of each of the samples was taken during fermentation on daily basis.
Sensory evaluation
Sensory evaluation of the fermented rice test samples were carried out with the use of questionnaires as described by Larmond [8]. Panelists were drawn from adults within Federal University of Technology, Akure, Nigeria. The samples were coded and presented to panellists to score for acceptability on appearance/colour, texture and aroma using a 9 point Hedonic scale.
Statistical analysis
Data obtained from the sensory evaluation were subjected to analysis of variance (ANOVA) and the means were separated with the aid of Duncan’s Multiple Range Test using SPSS 15.0 statistical software.
Daily occurrence of bacteria involved in the Igbemo rice sample fermentation was as shown in Table 1 below. Bacteria isolated from the rice sample were of the Bacillus and Erwinia genera throughout the 7- day fermentation period. At zero day, only B. cereus and B. subtilis (with 2.0 × 103 and 1.0 × 103 cfu/g respectively) were isolated. Their populations ranged between 1.0 × 103- 1.9 × 104 cfu/g and increased on daily basis till the fourth day with the highest populations of the two organisms. From the fifth day of fermentation, populations of the two bacteria started declining and reached their lowest populations (1.0 × 103 cfu/g) on the seventh day (Table 1). These findings are in agreement with report of Owens [9] that fermented and unfermented steamed glutinous rice normally supports the growth of Bacillus cereus to populations of up to 1.0 × 108 cfu/g and the organism populations’ increase during fermentation of tape-a rice recipe, without exceeding 1.0 × 105 cfu/g.
| Organisms (Bacteria) |
Days of fermentation/population (cfu/g) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| E. amylovora | - | 1.0 × 103 | 3.0 × 103 | - | - | - | - | - |
| E. tracheiphila | - | - |
|
- | - | - | - | |
| B. cereus | 2.0 × 103 | 5.0 × 103 | 8.0 × 103 | 1.2 × 104 | 1.9 × 104 | 5.0 × 103 | 3.0 × 103 | 1.0 × 103 |
| B. subtilis | 1.0 × 103 | 3.0 × 103 | 8.0 × 103 | 1.0 × 104 | 1.9 × 104 | 3.0 × 103 | 3.0 × 103 | 1.0 × 103 |
| B. megaterium | - | - | 5.0 × 103 | 8.0 × 103 | 2.0 × 103 | - | - | - |
| B. polymyxa | - | - | - | 3.0 × 103 | 3.0 × 103 | - | - | - |
| B. maceans | - | - | 4.0 × 104 | 2.0 × 103 | 1.0 × 103 | - | - | - |
| B. brevis | - | - | - | - | 4.0 × 103 | 3.0 × 103 | - | - |
Table 1: Populations of bacteria isolated from fermenting Igbemo rice.
However, B. megaterium (2.0 × 103-8.0 × 103 cfu/g) and B. maceans (1.0 × 103-4.0 × 104 cfu/g) were isolated from the sample on the second to fourth days of fermentation. Erwinia amylovora (1.0 × 103-3.0 ×103 cfu/g) was isolated on the first and second days and Bacillus polymyxa with fairly constant population was also isolated from the rice sample on the third and fourth days of fermentation, while B. brevis (3.0 × 103-4.0 × 103 cfu/g) was only isolated on the fourth and fifth days and E. tracheiphila was only isolated on the second day (Table 1).
Throughout the fermentation period, the highest bacteria population was recorded on the fourth day, while the least total population was obtained on the seventh day (Figure 1). The dominance of the freshly cooked and fermenting Igbemo rice bacteria population by members of the Bacillus spp . agreed with the findings of Cook et al. [10], which asserted that Bacillus spp . may occur (in fermentation) because their spores are commonly present in cereals and are able to survive the steaming (heat) treatment during processing.
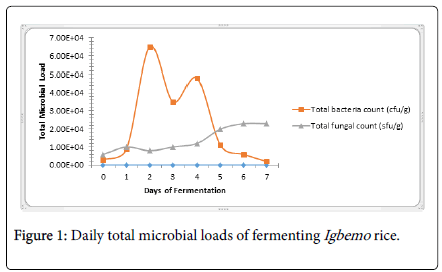
Figure 1. Daily total microbial loads of fermenting Igbemo rice.
The occurrence/populations of fungi isolated from Igbemo rice sample during the 7-day fermentation period was shown on Table 2. On days 0 and 1, four fungi species: Vigaria nigra , Penicillium italicum, Varicoporium elodeae and Articulospora inflata were isolated. Their populations ranged between 1.0 × 103-3.0 × 103 sfu/g of the rice sample, while V. nigra and P. italicum gave similar highest populations (3.0 × 103 sfu/g) on day 0. V. elodeae and A. inflata were of lower populations (1.0 × 103 sfu/g) respectively. Saccharomyces cerevisiae was however isolated on the second to the seventh day of fermentation (Table 2). The populations of S. cerevisiae isolated throughout the fermentation period under study ranged between 5.0 × 103-1.2 ×104 sfu/g. The organism had its highest and lowest populations (2.3 ×104 and 5.0 × 103 sfu/g) on the seventh and second days of fermentation respectively. The consistent isolation of Saccharomyces cerevisiae in this study was reported also in the findings of Suprianto et al. [11,12], which stated that Saccharomyces spp . were involved in the successful fermentation of rice for the production of Tapai in Asia.
| Organisms (Fungi) |
Days of fermentation/population (sfu/g) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Vigaria nigra | 2.0 × 103 | 3.0 × 103 | - | - | - | - | - | - |
| Penicillin italicum | 2.0 × 103 | 3.0 × 103 | 3 × 103 | 2.0 × 103 | - | - | - | - |
| Varicoporium Elodeae |
1.0 × 103 | 1.0 × 103 | - | - | - | - | - | - |
| Articulospora Inflate |
1.0 × 103 | 3.0 × 103 | - | - | - | - | - | - |
| Saccharomyces cerevisiae | - | - | 5.0 × 103 | 8.0 × 103 | 1.2 × 104 | 2.0 × 104 | 2.3 × 104 | 2.3 × 104 |
Table 2: Populations of fungi isolated from fermenting Igbemo rice.
Lowest populations of V. nigra (2.0 × 103 sfu/g), P. italicum (2.0 × 103 sfu/g), V. elodeae (1.0 × 103 sfu/g) and A. inflata (1.0 × 103 sfu/g) were obtained on the day 0 of fermentation. The population of V. elodeae however remained constant on the first day of fermentation, while the population of each of the other three fungi isolates increased from 1.0 × 103 sfu/g and 2.0 × 103 sfu/g to 3.0 × 103 sfu/g. Only P. italicum was isolated from the rice sample on the second and third days of fermentation out of the four fungi as shown in Table 2 above.
Daily populations of bacteria and fungi involved in the fermentation of the local Igbemo rice were shown in Figure 1. Occurrences of these organisms throughout the seven-day fermentation period ranged between 2.0 × 103-6.5 × 104 cfu/g. The bacteria population increased progressively from day 0 and reached its peak (6.5 × 104 cfu/g) on day 2. The population then dropped sharply on the third day to 3.5 ×104 cfu/g. The population increased again on the fourth day and dropped sharply to 1.1 × 104 cfu/g on the fifth day, after which the population began to drop steadily till the seventh day. The sudden drop in bacteria population may be due to accumulation of toxic wastes and continuous reduction in the environmental pH [13]. This was further explained by USDA’s [14] assertion that most bacteria are rapidly killed at pH and acid concentrations typical of acidified foods. The initial increase in the bacteria population till day 1 of fermentation could however be attributed to the fact that the few bacteria that survived cooking found the progression to anaerobic condition of fermentation as well as the nutrient-rich environment conducive for rapid growth and development [15]. Population growth pattern of the fungi isolates was slightly similar to that of the bacteria on the first two days of fermentation. The fungi population (mainly S. cerevisiae ) thereafter gradually increased from the second day of fermentation (8.0 ×103 sfu/g) till the sixth day (2.3 × 104 sfu/g) as the environmental pH reduced (Figure 1). This was in agreement with the findings of Arino [16] which stated that S. cerevisiae grows well over a wide range of pH, but grows better in acidic pH.
Changes in the pH of the fermented “Igbemo ” rice were shown in Figure 2. The fermentation proceeded to acidity from day 0 to day 6 and increased to near neutral (6.15) on day 7. The variations experienced in pH as fermentation proceeded is in agreement with the findings of Owens [9], which opined that the production of lactic acid during rice fermentation was capable of reducing the pH to as low as 4.0 (due to the virtual absence of buffering compounds in the food). This value was capable of increasing within 60 hours of fermentation, presumably as a result of the oxidation of the acid by the fermenting fungi [9]. Also, the proteolytic activity of Bacillus species involved in the rice sample fermentation might have increased the pH (on day 7) due to protein breakdown [17].
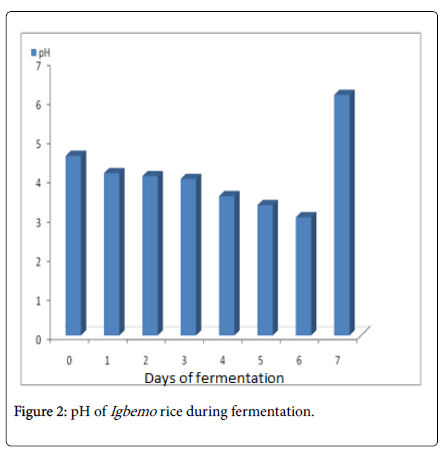
Figure 2. pH of Igbemo rice during fermentation.
Results of the sensory properties of the “Igbemo ” rice were shown in Table 3. There was no significant difference (p ≤ 0.05) in the colour of the fermented rice all through the seven-day fermentation period. The textures of the fermented rice for the first two days were significantly more acceptable (p ≤ 0.05) than those of the fermented rice obtained on days 3 to 6 of fermentation. This may be due to the liquefaction effect of the fermenting microbes involved as the days of fermentation progressed as reported by Suprianto et al. [9,11]. This may also be attributed to higher breaking strain (pressure) and lower breaking stress necessary to give a favourable chewy mouth-feel compared with non-fermented rice noodles [18].
| Days of Fermentation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Properties | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Texture | 7.9 ± 0.33a | 7.8 ± 0.29a | 7.7 ± 0.30a | 6.2 ± 0.36bc | 7.4 ± 0.37ab | 7.2 ± 0.55ab | 5.6 ± 0.64c | 7.5 ± 0.48ab |
| Colour | 7.7 ± 0.34abc | 8.2 ± 0.20a | 7.5 ± 0.22abc | 6.6 ± 0.31c | 7.5 ± 0.30abc | 7.3 ± 0.45abc | 6.7 ± 0.70bc | 8.0 ± 0.21a |
| Aroma | 7.1 ± 0.28a | 6.8 ± 0.25a | 6.7 ± 0.40a | 6.8 ± 0.36a | 7.4 ± 0.31a | 6.8 ± 0.44a | 6.1 ± 0.62ab | 6.8 ± 0.57a |
Values with the same subscripts along same row are not significantly different at P ≤ 0.05 using Duncan’s Multiple Range Test (DMRT)
Table 3: Effect of fermentation on the sensory properties of the fermented “igbemo” rice.
The appearance (colour) of the fermented rice obtained on day 7 was significantly more acceptable (p ≤ 0.05) than those obtained only on days 3 and 6 of fermentation, while those on other days were quite similar (Table 3). There was no significant difference (p ≤ 0.05) in the aroma of all the rice samples. Moreover, the unnoticeable changes in the rice aroma throughout the fermentation period were at variance with findings of Owens, Suprianto et al. [9,11].
Microbes involved in the fermentation of Igbemo rice were mainly members of the Bacillus and Saccharomyces genera. The fermented rice was most preferable and acceptable on the seventh day of fermentation with the least total microbial load. The pH of the sample at this stage was also at highest (near neutral), thus may be more ideal for consumption. Further research should however be conducted to determine the status of its nutrients as fermentation progresses and energy added value. This would help in knowing when best to consume the fermented food.
Citation: Babatuyi CY, Boboye BE, Ogundeji BA (2019) Dynamics of Microbial Populations on Fermented Nigerian ââ?¬Å?Igbemoââ?¬Â Rice in Akure, Nigeria. J Food Microbiol Saf Hyg 4:142.
Received: 15-Mar-2019 Accepted: 03-Apr-2019 Published: 10-Apr-2019 , DOI: 10.35248/2746-2059.20.5.142
Copyright: �© 2019 Babatuyi CY, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.